- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
An Assay to Study Botrytis cinerea-infected Grapevine Leaves Primed with Pseudomonas fluorescens
Published: Vol 6, Iss 19, Oct 5, 2016 DOI: 10.21769/BioProtoc.1943 Views: 12832
Reviewed by: Arsalan DaudiRupesh PaudyalAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Quantification of the Composition Dynamics of a Maize Root-associated Simplified Bacterial Community and Evaluation of Its Biological Control Effect
Ben Niu and Roberto Kolter
Jun 20, 2018 10488 Views

Tracking Root Interactions System (TRIS) Experiment and Quality Control
Hassan Massalha [...] Asaph Aharoni
Apr 20, 2019 7336 Views

A Quick Method for Screening Biocontrol Efficacy of Bacterial Isolates against Bacterial Wilt Pathogen Ralstonia solanacearum in Tomato
Heena Agarwal [...] Niraj Agarwala
Nov 20, 2020 5548 Views
Abstract
Grapevine (Vitis vinifera L.) is susceptible to an array of diseases among them the grey mold caused by the necrotrophic fungus Botrytis cinerea that decreases grape productivity and quality. To ensure a satisfactory yield and harvest quality numerous chemical fungicides are required, but they have serious drawbacks. One alternative is the use of beneficial bacteria to improve plant health. Pseudomonas fluorescens has been shown to trigger a plant-mediated resistance response in aboveground plant tissues against fungal, oomycete, bacterial, and viral pathogens. Triggered plant resistance exploits mechanisms of the plant immune system through a priming state that provides plants with enhanced capacity for rapid and strong activation of defense responses after pathogen infection, resulting in a lower fitness-cost. The primed responses by beneficial bacteria include induced expression of defense-related genes, cell wall reinforcement, and the production of secondary metabolites after pathogen infection. In this protocol, we describe the experimental design to evaluate the priming state of grapevine plants by the beneficial bacterium Pseudomonas fluorescens PTA-CT2 and their resistance level to Botrytis cinerea according to Verhagen et al. (2011) and Gruau et al. (2015).
Keywords: BiocontrolMaterials and Reagents
- 25 mm culture tubes (VWR, catalog number: 212-0304 )
- Conical tubes (50 ml) (Corning, Falcon®, catalog number: 352070 )
- Sterile spatula (VWR, catalog number: 612-1561 )
- Falcon® cell strainer 100 µm nylon (Corning, Falcon®, catalog number: 352360 )
- 1.5 ml microtubes (Treff, catalog number: 96.07246.9.01 )
- 2 ml microtubes (Treff, catalog number: 96.09329.9.01 )
- Whatman paper ashless discs 90 mm diameter (Sigma-Aldrich, catalog number: WHA1441090 )
- Petri dishes (90 x 14 mm) (VWR, catalog number: 391-0439 )
- Grapevine (Vitis vinifera cv. Chardonnay 7535) plants obtained by micropropagation from nodal explants on agar Murashige & Skoog (MS) medium (Aziz et al., 2006) in 25 mm culture tubes
- Pseudomonas fluorescens PTA-CT2 (Trotel-Aziz et al., 2008)
- Botrytis cinerea conidia (Bc 630) isolated from infected grape at ripening by INRA Versailles, France
- Distilled water from Aquadem blue range (Véolia Water Aquadem)
- Distilled water sterilized with autoclave Avor Getinge
- Ethanol (from Alcool absolu) (Charbonneaux Brabant, catalog number: 3077319001001 )
- Murashige & Skoog medium (MS) (Duchefa Biochimie, catalog number: M0231.0001 )
- Glycerol (VWR, catalog number: 24387-292 )
- Luria Bertani liquid medium (LB) (Duchefa Biochimie, catalog number: L1703.0500 )
- Magnesium sulfate heptahydrate (MgSO4·7H2O) (Sigma-Aldrich, catalog number: M2773 )
- BactoTM agar (BD, catalog number: 214010 )
- Agar tomato medium (see Recipes)
Equipment
- Plant growth chamber with photoperiod (SANYO, model: MLR-351 H )
- Horizontal laminar flow cabinet (ASTEC Microflow, model: Horizontal laminar flow workstation )
- Long sterile metal clamp (VWR, catalog number: 232-0084 )
- Magenta boxes (Sigma-Aldrich, catalog number: V8505 )
- Microbiological safety cabinet (Angelantoni Life Science, model: VBH Compact )
- Sterile pipettes (VWR, catalog number: VHPA23004 )
- 28 °C incubator shaker (New Brunswick Scientific, model: C24 )
- Variable speed refrigerated centrifuge with rotor having number of rotor cavities x nominal largest load (capacity: 8 x 50 ml) (Eppendorf, model: 5804R )
- Spectrophotometer (Bio-Rad Laboratories, model: SmartSpec 3000 )
- Vortex mixer (Vortex Genie 2 untimed mixer) (VWR, catalog number: 58815-232 )
- Malassez counting chamber (Marienfeld, catalog number: 0640610 )
- Glass atomizer (VWR, catalog number: 552-0031 )
- Camera (Nikon, model: D7000 )
Software
- APS Assess 2.0 software (APS, Item No: 43696M5, ISBN 978-0-89054-369-6)
Procedure
- Preparation of plant material
Grapevine vitro-plantlets (Vitis vinifera L. cv. Chardonnay clone 7535) were micropropagated from nodal explants grown on 15 ml of agar-modified Murashige-Skoog (MS) medium (with 0.7% of Bacto-agar) (Aziz et al., 2003) in 25 mm culture tubes (Figure 1).
Plants were grown at 25 °C with a 16- and 8-h photoperiod. Under sterile conditions (horizontal laminar flow cabinet) 8-week-old vitro-plantlets are gently removed from the MS agar and transferred with a long sterile metal clamp to sterile Magenta boxes containing sterile liquid MS medium, then closed for 24 h before treatment for acclimation. Each Magenta box contains four sterile lids filled with 9 ml of sterile liquid MS medium and the roots of each plant were immersed individually in each medium (Figure 1).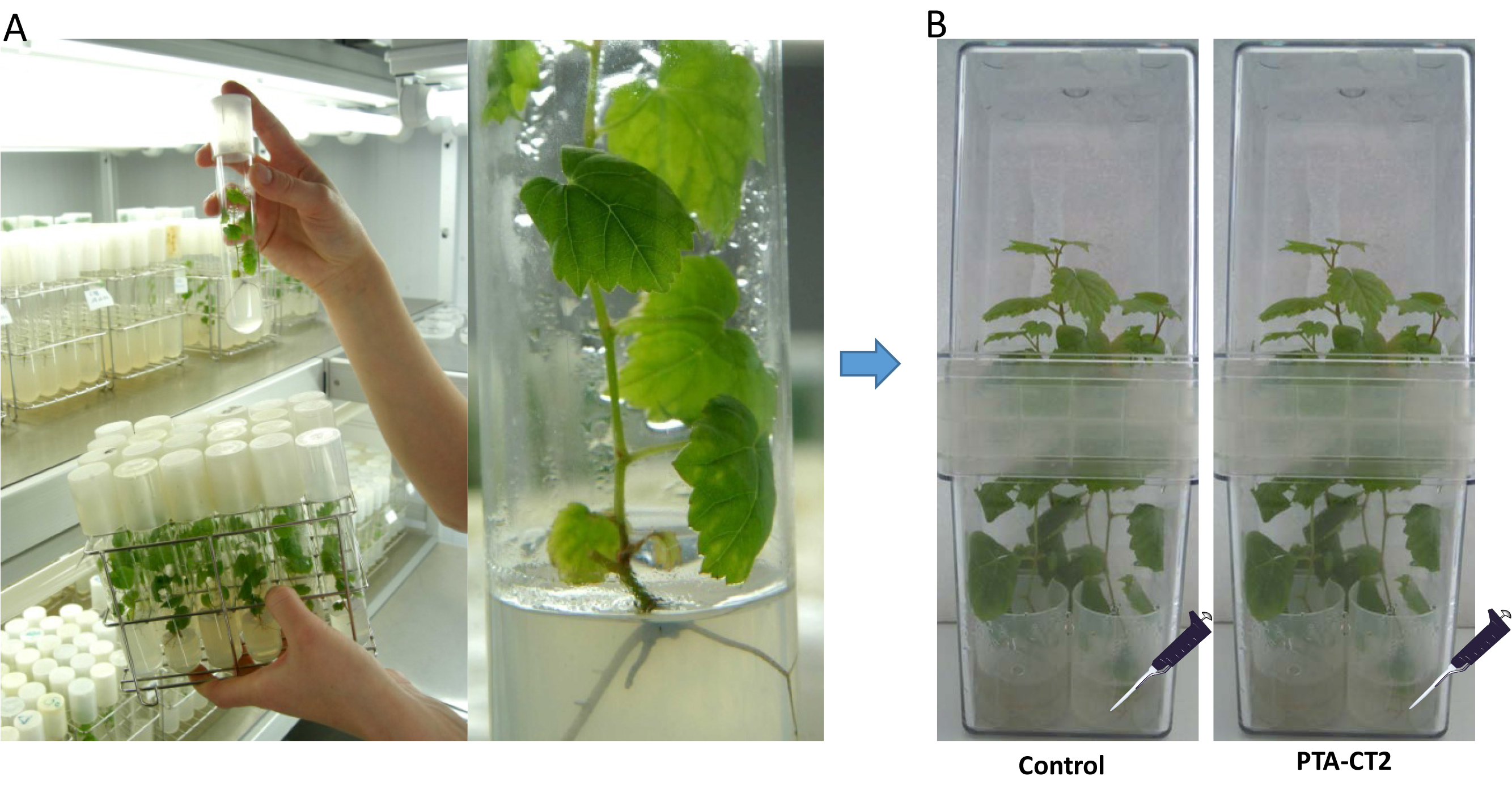
Figure 1. Procedure of inoculation with Pseudomonas fluorescens. A. Grapevine plants (Vitis vinifera L. cv. Chardonnay) were sensitive to Botrytis cinerea. B. Grapevine plants transferred to sterile Magenta boxes and inoculated with the bacterium Pseudomonas fluorescens PTA-CT2 at root level. - Bacterial inoculum preparation and inoculation
- Withdraw Pseudomonas fluorescens PTA-CT2 strain (Trotel-Aziz et al., 2008) from -80 °C glycerol stock (30%) and in the microbiological safety cabinet use a sterilized pipette tip to inoculate the bacterium in sterile Luria Bertani (LB) liquid medium (100 μl of glycerol stock for 10 ml of culture medium) and grow for 18 h in a 28 °C incubator shaker at 110 rpm.
- Use this preculture for 10 new LB cultures by inoculating 1 ml aliquots in 10 x 50 ml sterile tubes containing each 40 ml of LB under the same conditions as in paragraph B1.
- Centrifuge the bacterial suspension at 5,000 x g for 10 min, and in the microbiological safety cabinet wash the pellet with sterile 10 mM MgSO4 solution and resuspend it in 10 ml sterile 10 mM MgSO4 solution or MS medium.
- Vortex the cell suspension well and check its optical density at 450 and 600 nm (OD450 and OD600) by using a spectrophotometer. To prepare the inoculum, adjust the density of the bacterial suspension to 2 x 108 cfu/ml in 10 mM MgSO4 or in liquid MS medium.
- With a sterile pipet add 1 ml of bacterial suspension to the roots of each plantlet placed in lids in sterile conditions, to reach a final concentration of 2 x 107 cfu/ml (Figure 2A). For control plantlets, add 1 ml of 10 mM MgSO4 or of liquid MS medium.
- Place bacterized and control plantlets in a growth chamber at 22 °C with a 16- and 8-h photoperiod.
- For statistical analysis, prepare a minimum of twelve biological replicates.
- Withdraw Pseudomonas fluorescens PTA-CT2 strain (Trotel-Aziz et al., 2008) from -80 °C glycerol stock (30%) and in the microbiological safety cabinet use a sterilized pipette tip to inoculate the bacterium in sterile Luria Bertani (LB) liquid medium (100 μl of glycerol stock for 10 ml of culture medium) and grow for 18 h in a 28 °C incubator shaker at 110 rpm.
- Fungal inoculum preparation and inoculation
- Withdraw conidial suspension of Botrytis cinerea 630 from -80 °C glycerol stock (30%) and in the microbiological safety cabinet use a sterilized pipette tip to prick out conidia (10 µl) on a Petri dish containing tomato agar medium and incubate it for 21 days at 22 °C under continuous light for sporulation.
- Put 10 ml sterile water on sporulated B. cinerea and scrape with a sterile spatula to dislodge the conidia and mycelium of B. cinerea.
- Transfer the mixture of mycelium and conidia into the 50 ml conical tube, then shake it by vortexing several times to obtain conidia in water.
- Filter the obtained suspension using a filter tube (50 ml Falcon® tube containing 100 µM nylon Falcon® cell strainer) to remove mycelium.
- Measure the concentration of conidia in the suspension using a Malassez counting chamber and dilute with sterile water to a concentration of 1 x 106 conidia per ml water.
- For priming experiments and B. cinerea development assays, spray leaves of root-treated plants with P. fluorescens PTA-CT2 or control plants with the inoculation suspension of B. cinerea at 1 x 106 conidia per ml using an atomizer (Figure 2B), until homogenous coverage of the leaves is reached.
- Keep plants in a growth chamber at 22 °C with a 16- and 8-h photoperiod.
- Collect samples at 24 h post inoculation and grind the leaves with a mortar-pestle in liquid nitrogen to obtain a thin powder kept frozen.
- Weigh 50 mg of the frozen fresh leaf powder in a precooled 2 ml microtube before storage at -80 °C for further mRNA extraction and quantification of Actin gene expression of B. cinerea (EDN28275.1, GenBank) by qRT-PCR (Figure 2D) as described in Gruau et al. (2015).
- For the evaluation of long lasting resistance to B. cinerea, detach leaves from 5 day bacterized or control plants (Figure 2C).
- Put detached leaves with the upper face in contact with humidified Whatman paper discs in Petri dishes.
- Apply one needle-prick wound to the middle part of each leaf (Figure 2C), and cover the fresh wounds with 5-μl drops of a suspension of 1 x 106 conidia/ml.
- Keep Petri dishes containing leaves in a growth chamber at 22 °C with a 16- and 8-h photoperiod for 72 h post infection.
- Take pictures of the upper side of inoculated leaves with a Nikon camera D7000 and scan them, then measure the necrotic lesion areas formed during B. cinerea infection by Image Analysis Software, APS Assess 2.0 software (American Phytopathological Society Press, St. Paul, MN, USA) for necrosis surface quantification (Figure 2E).
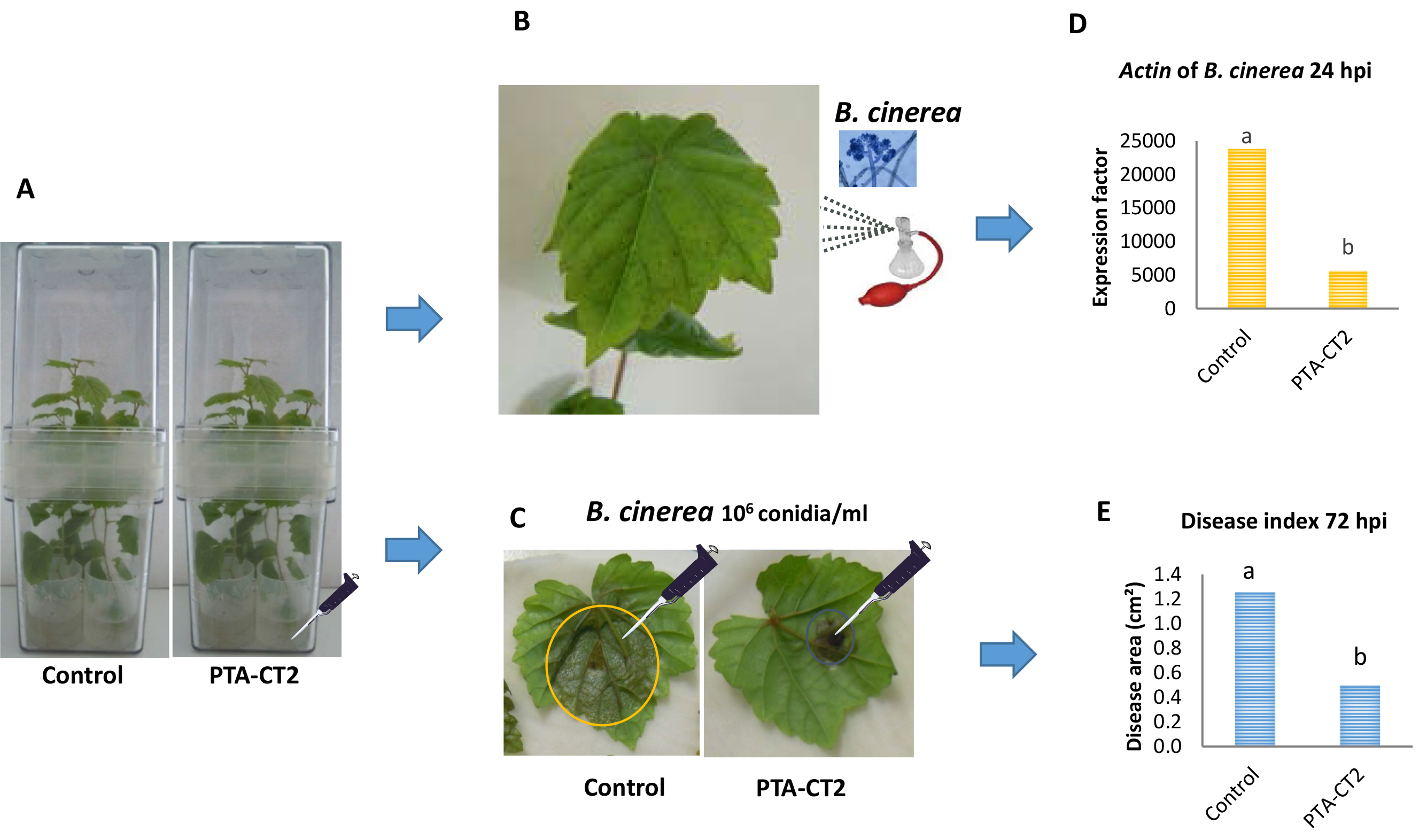
Figure 2. Procedure of priming plants with Pseudomonas fluorescens after challenge with B. cinerea. Five days post inoculation of grapevine plants with P. fluorescens PTA-CT2 in Magenta boxes at root level (A), leaves from control and treated plants were either sprayed with B. cinerea conidia (B), or detached and drop-inoculated with B. cinerea conidia (C). Evaluate the pathogen development during the first 24 hpi with the spray of B. cinerea conidia by quantifying the Actin transcript level of B. cinerea (as in D), or the mean necrosis area formed at 72 hpi in drop-infected leaves (as in E).
- Withdraw conidial suspension of Botrytis cinerea 630 from -80 °C glycerol stock (30%) and in the microbiological safety cabinet use a sterilized pipette tip to prick out conidia (10 µl) on a Petri dish containing tomato agar medium and incubate it for 21 days at 22 °C under continuous light for sporulation.
Data analysis
Constitutive Actin gene of B. cinerea in leaves of intact plants was analyzed by quantitative real-time RT-PCR using specific primers (see Grau et al., 2015). The reference genes EF1α (BQ799343, GenBank) and 60RSP (XM_002270599, GenBank) were used as internal controls and leaves of infected plantlets at zero time correspond to the reference sample (1x expression level). The results presented are means from three independent experiments.
Necrosis surface provoked by B. cinerea on detached leaves was acquired with the APS Assess 2.0 software. About 30 leaves were used for each condition in one experiment and the results are means of six independent experiments. ANOVA test was performed using Duncan’s multiple range test; P < 0.05.
Recipes
- Agar tomato medium
25% (v/v) commercial 100% tomato juice
2.4% (v/v) agar
Acknowledgments
This work was partially supported by the Vineal 2 program, and grants to C. Gruau and B. Verhagen from the Champagne-Ardenne Region and the City of Reims. This protocol was adapted from Gruau et al. (2015) and Verhagen et al. (2011).
References
- Aziz, A., Poinssot, B., Daire, X., Adrian, M., Bezier, A., Lambert, B., Joubert, J. M. and Pugin, A. (2003). Laminarin elicits defense responses in grapevine and induces protection against Botrytis cinerea and Plasmopara viticola. Mol Plant Microbe Interact 16(12): 1118-1128.
- Aziz, A., Trotel-Aziz, P., Dhuicq, L., Jeandet, P., Couderchet, M., and Vernet, G. (2006). Chitosan oligomers and copper sulfate induce grapevine defense reactions and resistance to gray mold and downy mildew. Phytopathology 96 (11):1188-1194.
- Gruau, C., Trotel-Aziz, P., Villaume, S., Rabenoelina, F., Clement, C., Baillieul, F. and Aziz, A. (2015). Pseudomonas fluorescens PTA-CT2 triggers local and systemic immune response against Botrytis cinerea in grapevine. Mol Plant Microbe Interac 28(10): 1117-1129.
- Trotel-Aziz, P., Couderchet, M., Biagianti, S. and Aziz, A. (2008). Characterization of new bacterial biocontrol agents Acinetobacter, Bacillus, Pantoea and Pseudomonas spp. mediating grapevine resistance against Botrytis cinerea. Environ and Exp Bot 64(1): 21-32.
- Verhagen, B., Trotel-Aziz, P., Jeandet, P., Baillieul, F. and Aziz, A. (2011). Improved resistance against Botrytis cinerea by grapevine-associated bacteria that induce a prime oxidative burst and phytoalexin production. Phytopathology 101(7): 768-777.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Gruau, C., Trotel-Aziz, P., Verhagen, B., Villaume, S., Rabenoelina, F., Courteaux, B., Clément, C., Baillieul, F. and Aziz, A. (2016). An Assay to Study Botrytis cinerea-infected Grapevine Leaves Primed with Pseudomonas fluorescens . Bio-protocol 6(19): e1943. DOI: 10.21769/BioProtoc.1943.
Category
Plant Science > Plant immunity > Disease bioassay
Plant Science > Plant immunity > Perception and signaling
Microbiology > Microbe-host interactions > In vivo model > Plant
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link














