- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Extraction and Quantification of GABA and Glutamate from Cyanobacterium Synechocystis sp. PCC 6803
Published: Vol 6, Iss 18, Sep 20, 2016 DOI: 10.21769/BioProtoc.1928 Views: 12439
Reviewed by: Maria SinetovaSeda EkiciVinai Chittezham Thomas

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

β-lactamase (Bla) Reporter-based System to Study Flagellar Type 3 Secretion in Salmonella
Fabienne F. V. Chevance and Kelly T. Hughes
Jun 20, 2023 1765 Views

Determination of Poly(3-hydroxybutyrate) Content in Cyanobacterium Synechocystis sp. PCC 6803 Using Acid Hydrolysis Followed by High-performance Liquid Chromatography
Janine Kaewbai-ngam [...] Tanakarn Monshupanee
Aug 20, 2023 1809 Views

An HPLC-based Assay to Study the Activity of Cyclic Diadenosine Monophosphate (C-di-AMP) Synthase DisA from Mycobacterium smegmatis
Avisek Mahapa [...] Dipankar Chatterji
Dec 20, 2024 1780 Views
Abstract
GABA (γ-amino butyric acid) is a biologically active four carbon non-protein amino acid found in prokaryotes and eukaryotes. Glutamate is a five carbon α-amino acid which can be converted to GABA catalyzed by the enzyme glutamate decarboxylase. In this protocol, we describe the procedure for extraction and quantification of GABA and glutamate from cells of the cyanobacterium Synechocystis sp. PCC 6803 (hereafter Synechocystis). Apart from Synechocystis, this protocol has already been successfully tested for the cyanobacterium Nostoc punctiforme and for the green alga Tetraspora sp. CU2551. The protocol can also be used for the analyses of other primary amino acids. We have successfully employed this protocol in our studies of GABA and glutamate analyses in Synechocystis (Kanwal and Incharoensakdi, 2016).
Keywords: GABA assayBackground
Amino acid analysis has been done previously by employing a number of methods that include the detection using thin layer chromatography or ion exchange separation coupled with post-column derivatization with ninhydrin method or also using HPLC by fluorescent detection. However, several challenges are encountered in those detection methods such as poor sensitivity, degradation of derivatives, formation of multiple derivatives and longer retention times causing higher solvent consumption. Furthermore, the UV detection methods alone could not be used owing to optically inactive nature of amino acids.
Here, we describe the detailed procedure for extraction and analyses of amino acids such as GABA and glutamate in Synechocystis. The method used for amino acids separation and detection is high performance reversed-phase liquid chromatography followed by UV detection of pre-column OPA (o-phthaldehyde) derivatization of amino acids adopted from Henderson et al. (2006). This method offers several advantages over previous methods by improving the sensitivity of detection. Reversed-phase chromatography allows faster separation of amino acids, whereas UV detection offers improved sensitivity. OPA derivatization of amino acids results in stable derivatives that make the UV detection of amino acids possible. The method also offers the detection and quantification of amino acids within 26 min, which consequently reduces solvent consumption.
Materials and Reagents
- 50 ml polypropylene tubes (Corning®, CentristarTM, Mexico)
- 1.5 ml Eppendorf tubes (Hycon plastics, catalog number: HCE-1004-MIC )
- 200 µl conical inserts with polymer feet (Borosilicate glass) (JG Finneran, USA)
- Amber wide-opening vial with screw cap (Borosilicate glass) (JG Finneran, USA)
- 0.2 µm Millipore filter (Nylon or cellulose filter membrane) (Sartorius Stedim biotech, Germany)
- Synechocystis cells (fresh or frozen at -20 °C)
- BG11 growth medium
- Deionized water
- Methanol, AR grade (RCI Labscan, Thailand)
- Ethanol, AR grade (QRec, New Zealand)
- Chloroform (RCI Labscan, Thailand)
- Hydrochloric acid (HCl) (QRec, New Zealand)
- NaH2PO4 (Carlo Erba Reagents, Chaussée du Vexin, Val de Reuil, France)
- NaOH (Merck, Germany)
- UP water, HPLC grade (TEDIA, OH, USA)
- Acetonitrile CAN, HPLC grade (Burdick and Jackson, Korea)
- Methanol (MeOH), HPLC grade (Burdick and Jackson, Korea)
- Standards of glutamate and GABA, > 99% purity (Sigma-Aldrich)
- 0.4 M borate buffer, pH 10.2 (Agilent Technologies, catalog number: 5061-3339 )
- O-phthaldehyde reagent (OPA) (Agilent Technologies, catalog number: 5061-3335 )
- BG11 growth medium (see Recipes)
- HPLC solvent A (see Recipes)
- HPLC solvent B (see Recipes)
- OPA reagent (see Recipes)
Equipment
- Pyrex® 250 ml conical flasks (Corning, Pyrex®, catalog number: 4980 )
- Shaker (Eppendorf, model: InnovaTM 2100 )
- Tabletop centrifuge (Hettich Lab Technology, model: MIKRO220/220R )
- Vortex mixer (Scientific Industries, model: SI-0236 )
- Ultrasonic laboratory homogenizer (Sonuplus UW2200)
- Centrivap concentrator (Labconco, USA)
- Shimadzu prominence ultra-fast liquid chromatography system (Shimadzu Scientific Instruments)
- Shimadzu prominence autosampler (Shimadzu Scientific Instruments, model: SIL-21A HT )
- Degasser (Shimadzu Scientific Instruments, model: DGU-20A5 )
- UV-vis detector (Shimadzu Scientific Instruments, model: SPD-20A )
- Reverse phase column (ZORBAX Eclipse AAA analytical column, 4.6 x 150 mm, 5.0 μm) (Agilent Technologies, catalog number: 93400-902 )
- Guard column (ZORBAX Eclipse AAA analytical guard column, 4.6 x 12.5 mm, 5.0 μm) (Agilent Technologies, catalog number: 820950-931 )
Procedure
- GABA and glutamate extraction
- Grow Synechocystis cells, at 30 °C in BG11 medium with a total volume of 50 ml in a cotton-plugged 250 ml conical flask.
- Harvest cells at exponential phase of growth (3 to 8 days) by centrifuging at 2,500 x g for 10 min, room temperature. Discard supernatant, resuspend cell pellet in 500 µl of deionized H2O and transfer the cell suspension into a 1.5 ml Eppendorf tube. Spin down the cells at 10,000 x g for 10 min and remove all water by micropipette (at this point, cells can be frozen at -20 °C for up to a month).
- Resuspend the cells in 1.5 ml of 80% (v/v) methanol by vigorous mixing. Incubate the mixture for a few hours at room temperature (or incubate overnight) and homogenize using ultrasonic homogenizer for 2 min.
- Pellet cell debris by centrifuging at 10,000 x g for 10 min, 4 °C. Collect and transfer the supernatant into a new 1.5 ml Eppendorf tube to obtain the “methanol extract”.
- Dry the methanol extract in a vacuum evaporator at 40 °C for 5 h and keep the dried pellet at room temperature (steps A6 and 7 can be omitted, see Note 1 below).
- To the cell debris obtained from step A4, add 2 ml of 80% (v/v) ethanol for further re-extraction in the same way to get the “ethanol extract” (see steps A2 and A3).
- Add the ethanol extract to previously obtained dried pellet from methanol extract at step A5 and dry in a vacuum evaporator at 40 °C.
- Dissolve the pellet obtained after drying with 250 µl of deionized water and 50 µl of chloroform, followed by centrifugation at 10,000 x g for 10 min, 4 °C. Collect the upper aqueous phase (collect sufficient amount of aqueous phase and be careful not to collect the chloroform phase) and transfer into a new 1.5 ml Eppendorf tube.
- Add diluted HCl to the aqueous phase to make a final concentration of 0.1 N. Filter the solution through a 0.2 µm Millipore filter before GABA/glutamate detection by HPLC. The filtered sample can be stored at -20 °C until further use.
- Grow Synechocystis cells, at 30 °C in BG11 medium with a total volume of 50 ml in a cotton-plugged 250 ml conical flask.
- HPLC analysis of GABA and glutamate
- Filter deionized water (or use UP water, HPLC grade), solvent A and solvent B through a 0.2 µm Millipore filter.
- The pre-column sample derivatization with OPA reagent is performed as described below.
- Transfer 30 µl of filtered sample from step A9 into a 1.5 ml Eppendorf tube and add 5 µl of borate buffer.
- Mix thoroughly and incubate for 15 sec at room temperature.
- Add 0.5 µl of OPA reagent and mix well (see Note 2).
- Transfer the solution into a 200 µl conical insert (Figure 1A). Place the insert in a transparent wide-opening vial (Figure 1B).
- Close the vial with screw cap (Figure 1C) and the sample is immediately injected into the HPLC system by an autoinjector.
- Separation is performed on a reverse phase column with solvent A and solvent B in 26 min at a flow rate of 2 ml/min at 40 °C (column oven temperature) using a gradient program (Table 1) (see Note 3). The UV-detection wavelength is 338 nm for OPA derivatized samples.
- Quantification of GABA and glutamate content is done by comparing the peak areas with those of standards (linearity is obtained for the concentration range of 4.5 to 450 pmol in 0.5 µl of sample). Retention time for glutamate and GABA using these HPLC conditions is 3.0-3.7 min and 10.0-10.5 min respectively (see the Data analysis, Figure 2) (see Note 4).

Figure 1. A photograph of (A) conical insert, (B) transparent wide-opening vial, and (C) screw cap for GABA and glutamate analyses using the Shimadzu prominence autosampler
Table 1. HPLC gradient program settings for a single run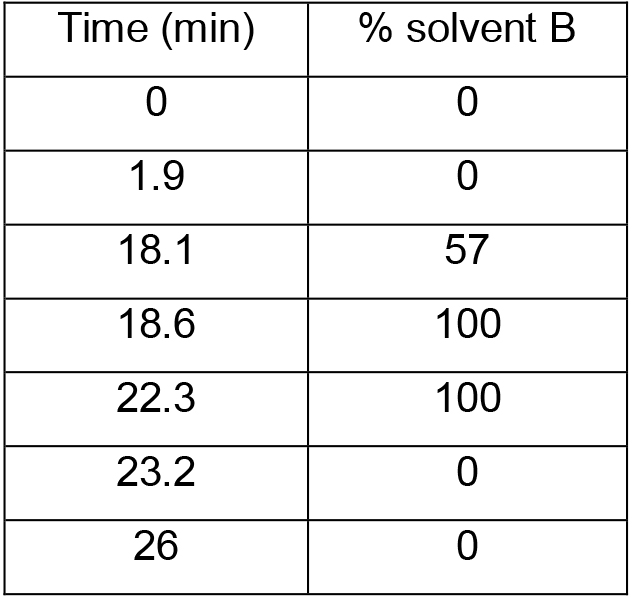
- Filter deionized water (or use UP water, HPLC grade), solvent A and solvent B through a 0.2 µm Millipore filter.
Data analysis
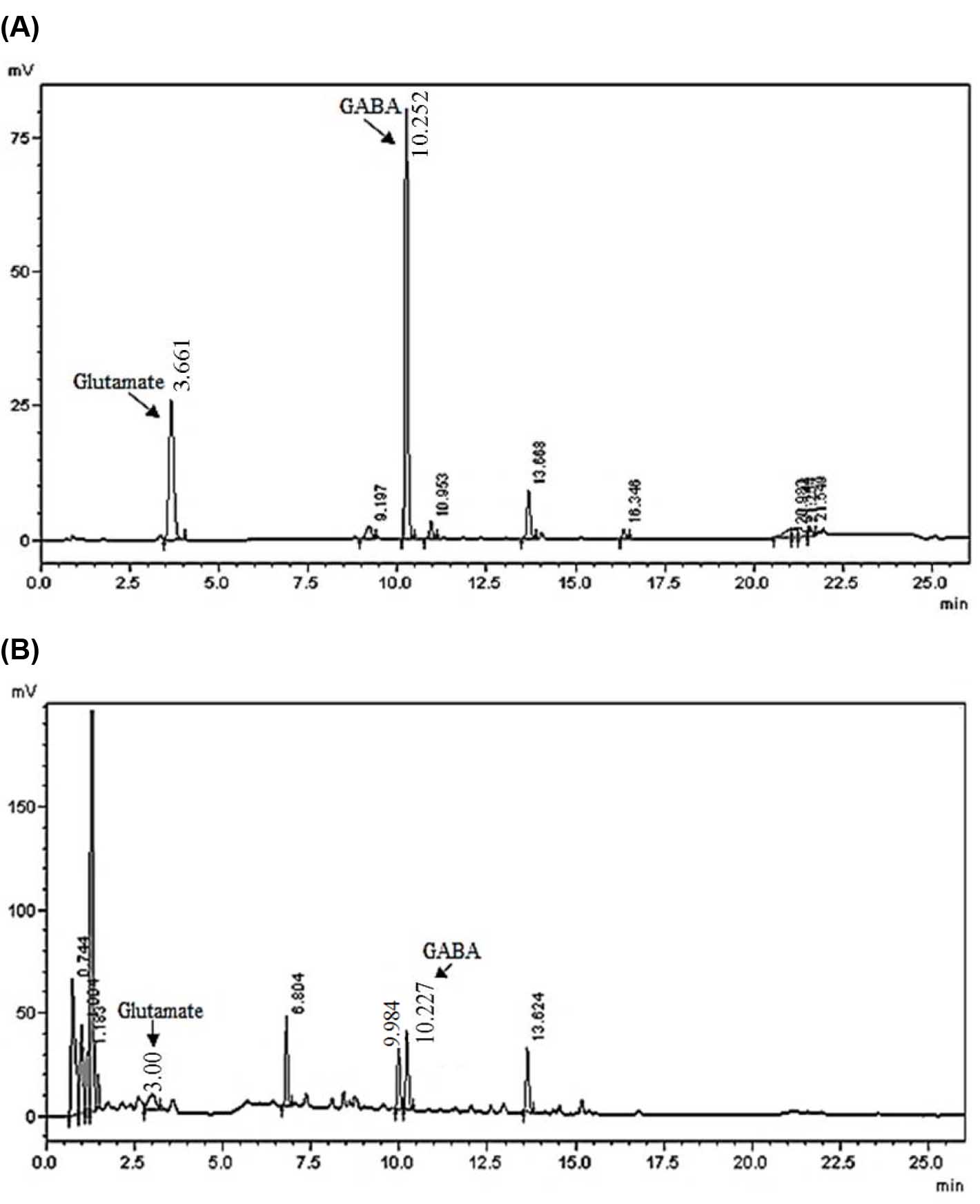
Figure 2. Representative chromatograms of separated amino acids extracted from the cyanobacterium Synechocystis sp. PCC 6803. A. Chromatogram of standard glutamate and GABA for the concentration of 2 nmol/ml; B. Chromatogram of amino acid separation from Synechocystis sp. PCC 6803 extract. Data were obtained from Shimadzu LC solution software and were confirmed by three independent experiments.
Notes
- The ethanol extraction step can be omitted. However, to ensure the complete quantitative recovery of amino acids, this step is recommended.
- The OPA reagent slowly degrades in the presence of oxygen. Inject immediately the freshly derivatized reaction mixture into HPLC.
- In case if further cleaning is required for column, the program could be continued by column washing with 100% solvent B for 20-30 min followed by equilibration with buffer (solvent A) for 20-30 min.
- To ensure the consistency of HPLC conditions and retention time of standards and sample, it is important to run standards regularly.
Recipes
- BG11 growth medium
Note: Preparation of BG11 growth medium is described in detail in Rippka et al., 1979. - HPLC solvent A
40 mM NaH2PO4, pH 7.8
Dissolve 5.5 g NaH2PO4·H2O in 1 L of water
Adjust pH with 10 N NaOH solution - HPLC solvent B
ACN:MeOH:water (45:45:10, v/v/v) - OPA reagent
10 mg/ml o-phthalaldehyde
10 mg/ml 3-mercaptopropionic acid
0.4 M borate buffer, pH 10.2
Acknowledgments
The method for GABA and glutamate extraction was adapted from Bieleski and Turner (1966) with some modifications. The HPLC-based method for the detection and quantification of GABA and glutamate is a modification of protocol published by Henderson et al. (2006). This work was supported by research grants from Chulalongkorn University (CU) Ratchadaphiseksomphot Endowment Fund (Food and Water Cluster, CU-58-011-FW) and from the Thailand Research Fund (IRG5780008). Simab Kanwal thanks CU Graduate School for providing post-doctoral fellowship.
References
- Bieleski, R. L. and Turner, N. A. (1966). Separation and estimation of amino acids in crude plant extracts by thin-layer electrophoresis and chromatography. Anal Biochem 17: 278-293.
- Henderson, J. W., Ricker, R. D., Bidlingmeyer, B. A. and Woodward, C. (2006). Rapid, accurate, sensitive, and reproducible HPLC analysis of amino acids. Agilent Application Note 5980-1193EN: 1-10.
- Kanwal, S. and Incharoensakdi, A. (2016). Characterization of glutamate decarboxylase from Synechocystis sp. PCC 6803 and its role in nitrogen metabolism. Plant Physiol Biochem 99: 59-65.
- Rippka, R., Deruelles, J., Waterbury, J. B., Herdman, M. and Stanier, R. Y. (1979). Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol 111: 1-61.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Kanwal, S. and Incharoensakdi, A. (2016). Extraction and Quantification of GABA and Glutamate from Cyanobacterium Synechocystis sp. PCC 6803. Bio-protocol 6(18): e1928. DOI: 10.21769/BioProtoc.1928.
Category
Microbiology > Microbial biochemistry > Other compound
Biochemistry > Other compound > Amino acid
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











