- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Analysis of Enteric Neural Crest Cell Migration Using Heterotopic Grafts of Embryonic Guts
Published: Vol 6, Iss 17, Sep 5, 2016 DOI: 10.21769/BioProtoc.1924 Views: 8509
Reviewed by: Jia LiGuillermo GomezJyotiska Chaudhuri

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Conditioned Lick Suppression: Assessing Contextual, Cued, and Context-cue Compound Fear Responses Independently of Locomotor Activity in Mice
Youcef Bouchekioua [...] Yu Ohmura
Dec 5, 2022 1641 Views

In situ Microinflammation Detection Using Gold Nanoclusters and a Tissue-clearing Method
Fayrouz Naim [...] Masaaki Murakami
Apr 5, 2023 2670 Views

A One-Step Mouse Model of Parkinson’s Disease Combining rAAV-α-Synuclein and Preformed Fibrils of α-Synuclein
Santhosh Kumar Subramanya [...] Poonam Thakur
Dec 5, 2025 1644 Views
Abstract
Hirschsprung disease (HSCR), also named aganglionic megacolon, is a severe congenital malformation characterized by a lack of enteric nervous system (ENS) in the terminal regions of the bowel (Bergeron et al., 2013). As the ENS notably regulates motility in the whole gastrointestinal track, the segment without neurons remains tonically contracted, resulting in functional intestinal obstruction and accumulation of fecal material (megacolon). HSCR occurs when enteric neural progenitors of vagal neural crest origin fail to fully colonize the developing intestines. These “enteric” neural crest cells (ENCCs) have to migrate in a rostro-caudal direction during a fixed temporal window, which is between embryonic day (e) 9.5 and e14.5 in the mouse (Obermayr et al., 2013). Recently, our group generated a new HSCR mouse model called Holstein in which migration of ENCCs is impaired because of increased collagen VI levels in their microenvironment (Soret et al., 2015). Here, we describe the method that allowed us to demonstrate the cell-autonomous nature of this migration defect. In this system adapted from a previously described heterotopic grafting approach (Breau et al., 2006), the donor tissue is a fully colonized segment of e12.5 midgut while the host tissue is an aneural segment of e12.5 hindgut. Extent of ENCC migration in host tissue is assessed after 24 h of culture and is greatly facilitated when donor tissue has a transgenic background such as the Gata4-RFP (Pilon et al., 2008) that allows endogenous labeling of ENCCs with fluorescence. Depending of the genetic background of donor and host tissues, this approach can allow evaluating both cell-autonomous and non-cell-autonomous defects of ENCC migration.
Keywords: Enteric nervous systemMaterials and Reagents
- Petri dishes (Corning, catalog number: 70165-102 )
- 24-well plate
- Nitrocellulose filter (Merck Millipore, catalog number: GSWP01300 )
- 8-chamber slides (ibiTreat μ-slide) (ibidi GmbH, catalog number: 80826 )
- Mature mice (≥ 2-month old)
- Isoflurane for inhaled anesthesia (Henry Schein Animal Health, catalog number: 050031 )
- 70% ethanol
- 1x phosphate-buffered saline (PBS)
- DMEM/Ham’s F-12 (WISENT, catalog number: 319-085-CL )
- Fetal Bovine Serum (WISENT, catalog number: 920-040 )
- Penicillin/streptomycin (WISENT, catalog number: 450-201-EL )
Equipment
- Dumont #5 dissection forceps (Fine Science Tools, catalog number: 11251-20 )
- Dumont #7 dissection forceps (Fine Science Tools, catalog number: 11274-20 )
- Dissection scissors (Moria Spring Scissors) (Fine Science Tools, catalog number: 15396-01 )
- Dissecting stereomicroscope (Leica Microsystems, model: M125 )
- CO2 cell culture incubator (Sanyo Scientific, model: MCO-18AIC )
Note: This product has been discontinued. - Infinity-2 camera (Lumenera Corporation) mounted on a fluorescent stereomicroscope (Leica Microsystems, model: M205FA )
Software
- ImageJ software
Procedure
- Mate mature mice (≥ 2-month old) overnight and check for the presence of a vaginal plug the next morning. Noon of the day a vaginal plug is observed is considered embryonic day (e) 0.5. A typical experiment requires one wildtype couple and one couple bearing the mutation to study.
- Twelve days later, euthanize isoflurane-anesthetized pregnant female(s) via CO2 inhalation.
- Use 70% ethanol to spray the mouse abdomen and open it with dissecting scissors to access the uterus.
- Remove the uterus into a glass petri dish containing 15 ml of sterile ice-cold PBS and cut it between individual embryos.
- Working on each embryo separately in another glass petri dish filled with 15 ml of ice-cold PBS, use fine forceps to remove the uterine muscle layers under a dissecting microscope (Figure 1 - step 1 and Video 1).
- Open the extra-embryonic membranes to access the embryo by taking care not to sever the developing intestines that are intertwined with the blood vessels connecting the embryo to the placenta/extra-embryonic membranes (Figure 1 - step 2 and Video 1).
- Cut embryo's head which can be used for PCR-based genotyping if necessary and open the abdominal cavity (Figure 1 - steps 3-4 and Video 1).
- After removing the liver, cut the esophagus and pull the whole gastrointestinal tract out of the abdominal cavity while being careful not to damage the colon still attached to the anus (Figure 1 - step 5 and Video 1).
- Cut at the anus to free the gastrointestinal tract. The stomach and cecum will allow keeping track of sample's orientation (Figure 1 - step 6 and Video 1).
Video 1. Whole dissection procedure - Transfer each sample into a well of a 24-well plate containing 1 ml of sterile PBS and kept on ice. If PCR genotyping is required, be careful to match intestine and head numbers for each sample.
- Assemble each graft by depositing and juxtaposing donor (wildtype or mutant) and host (wildtype or mutant) tissues onto a small 13 mm nitrocellulose membrane in a glass petri dish containing 15 ml of sterile ice-cold PBS. Prepare each half of a graft in a sequential manner, so that the first half has already adhered to the filter before processing the second half. Take care to respect the normal rostro-caudal orientation (esophagus to anus) of both the donor and host tissues in the assembled graft. Host tissue from a selected sample is prepared by cutting a ~0.5 cm section of the most caudal hindgut whereas donor tissue is prepared from another sample by cutting a ~0.5 cm section of the midgut just upstream of the cecum.
- Transfer each graft-bearing filter in a chamber of an 8-chamber slide containing 250 µl of DMEM/F12 supplemented with 10% FBS and penicillin/streptomycin (Figure 2A).
- After 24 h of culture (37 °C, 5% CO2), colonization of aneural hindgut tissues by midgut-derived ENCCs is quantified by measuring the distance separating the most distal ENCC from the midgut-hindgut graft junction (Figure 2B).
Representative data
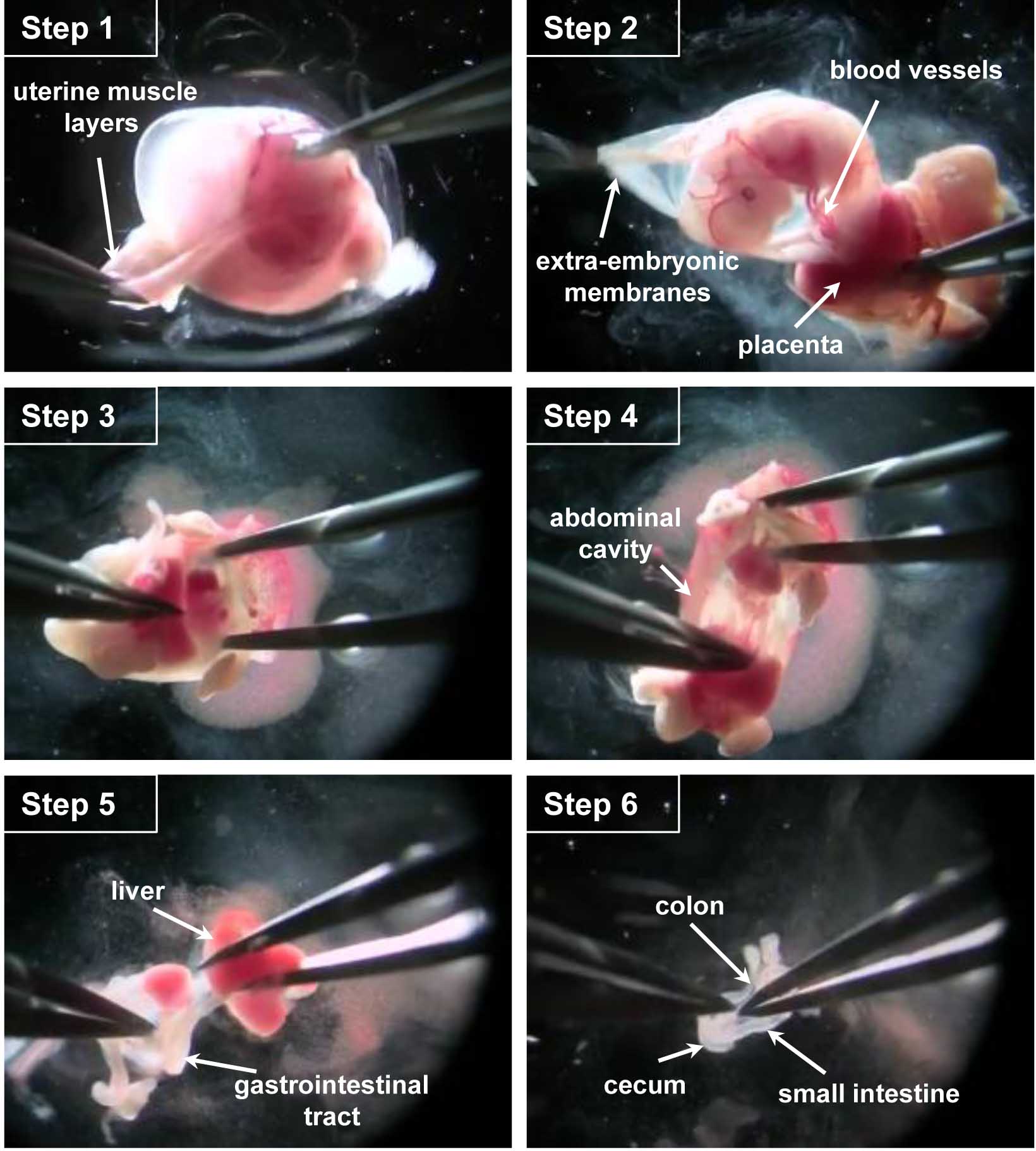
Figure 1. Key steps of the dissection procedure. Step 1. With fine forceps, remove the uterine muscle layers. Step 2. Open the extra-embryonic membrane to access the embryo, cut the blood vessels connecting the embryo to the placenta/extra-embryonic membranes. Step 3. Cut the embryo head and open the abdominal cavity. Step 4. Pull the whole gastrointestinal tract out of the abdominal cavity. Step 5. Remove the liver, cut the esophagus and release the digestive tract of the connective tissue. Step 6. Free the gastrointestinal tract by cutting it at the anus.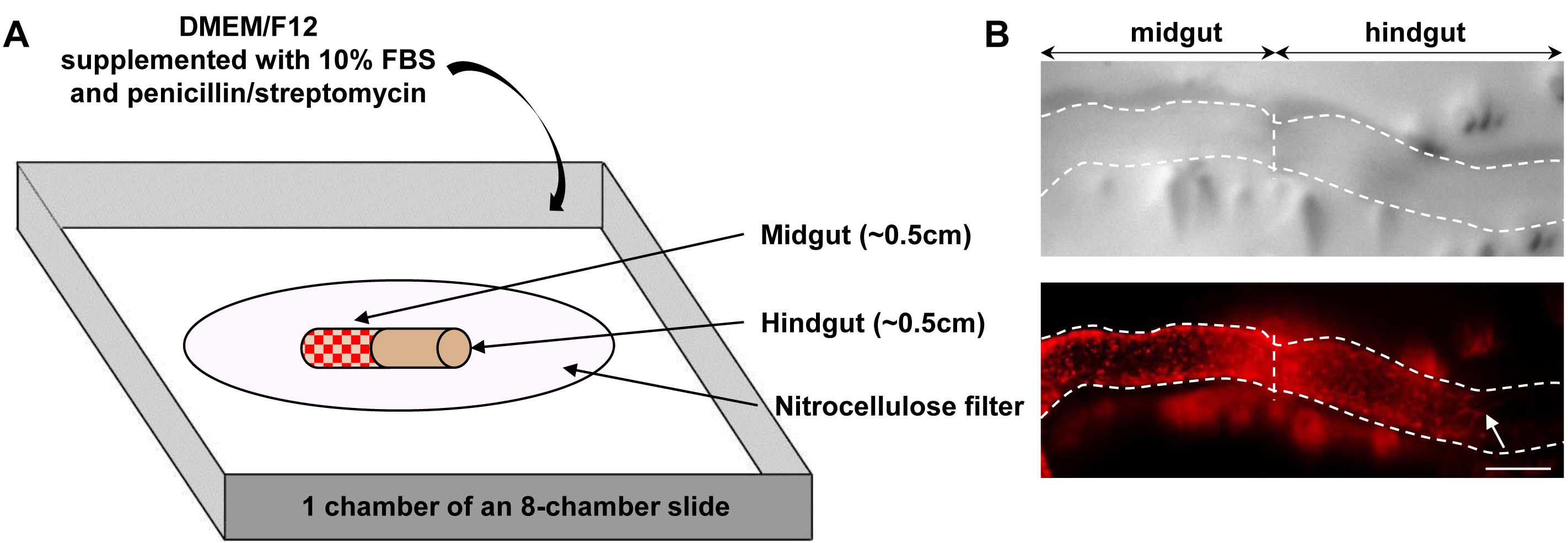
Figure 2. Overview of graft assembly and representative result. A. Schematic representation of a heterotopic e12.5 midgut-hindgut graft onto a nitrocellulose filter in a chamber of an 8-chamber slide. Red dots in midgut tissue represent ENCCs labeled by RFP (DsRed2) fluorescence provided by the G4-RFP transgene; B. Representative images of a graft (delineated by dotted lines) after 24 h of culture, showing the colonization of a previously aneural hindgut host tissue by fluorescently labeled ENCCs from midgut donor tissue. Pictures were taken using an Infinity-2 camera mounted on a Leica M205FA fluorescent stereomicroscope and images were analyzed using the ImageJ software. The white arrow points to the location of the migration front at the end of the culture period. Scale bar: 150 μm.
Notes
It is noteworthy that this method is greatly simplified when donor tissues are taken from embryos bearing a transgene such as the G4-RFP transgene (Pilon et al., 2008) that labels ENCCs with fluorescence. At step 10, this can allow the identification of mutant tissues (i.e., displaying delayed migration) by simple fluorescent microscopy instead of having to wait for genotyping results after graft assembly. Moreover, at step 13, such an intrinsic fluorescent labeling greatly facilitates the analysis of ENCC migration which otherwise requires immunofluorescence labeling using an antibody against a marker of undifferentiated enteric neural progenitors such as Sox10.
Acknowledgments
The Pilon laboratory is funded by grants from the Canadian Institute of Health Research (CIHR), the Natural Science and Engineering Research Council of Canada (NSERC) and the Fondation du grand défi Pierre Lavoie. RS holds a fellowship from the Fonds de la recherche du Québec Santé (FRQS) whereas NP is a FRQS Junior 2 Research Scholar as well as the recipient of a UQAM Research Chair on Rare Genetic Diseases. The authors thank the innovative work performed by Breau et al. (2006), on which this protocol was based.
References
- Bergeron, K. F., Silversides, D. W. and Pilon, N. (2013). The developmental genetics of Hirschsprung's disease. Clin Genet 83(1): 15-22.
- Breau, M. A., Pietri, T., Eder, O., Blanche, M., Brakebusch, C., Fassler, R., Thiery, J. P. and Dufour, S. (2006). Lack of β1 integrins in enteric neural crest cells leads to a Hirschsprung-like phenotype. Development 133(9): 1725-1734.
- Obermayr, F., Hotta, R., Enomoto, H. and Young, H. M. (2013). Development and developmental disorders of the enteric nervous system. Nat Rev Gastroenterol Hepatol 10(1): 43-57.
- Pilon, N., Raiwet, D., Viger, R. S. and Silversides, D. W. (2008). Novel pre- and post-gastrulation expression of Gata4 within cells of the inner cell mass and migratory neural crest cells. Dev Dyn 237(4): 1133-1143.
- Soret, R., Mennetrey, M., Bergeron, K. F., Dariel, A., Neunlist, M., Grunder, F., Faure, C., Silversides, D. W., Pilon, N. and Ente-Hirsch Study, G. (2015). A collagen VI-dependent pathogenic mechanism for Hirschsprung's disease. J Clin Invest 125(12): 4483-4496.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Soret, R. and Pilon, N. (2016). Analysis of Enteric Neural Crest Cell Migration Using Heterotopic Grafts of Embryonic Guts. Bio-protocol 6(17): e1924. DOI: 10.21769/BioProtoc.1924.
Category
Neuroscience > Nervous system disorders > Animal model
Developmental Biology > Cell growth and fate > Neuron
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











