- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Analytical Gel Filtration for Probing Heavy Metal Transfer between Proteins
Published: Vol 6, Iss 15, Aug 5, 2016 DOI: 10.21769/BioProtoc.1888 Views: 11103
Reviewed by: Valentine V TrotterEsteban Paredes-OssesAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Assay for Assessing Mucin Binding to Bacteria and Bacterial Proteins
Lubov S. Grigoryeva [...] Nicholas P. Cianciotto
Mar 5, 2021 4767 Views

Binding Affinity Quantifications of the Bacteriophage Mu DNA Modification Protein Mom Using Microscale Thermophoresis (MST)
Shubha Udupa [...] Shweta Karambelkar
Jul 20, 2022 3699 Views

Large-scale Purification of Type III Toxin-antitoxin Ribonucleoprotein Complex and its Components from Escherichia coli for Biophysical Studies
Parthasarathy Manikandan [...] Mahavir Singh
Jul 5, 2023 2169 Views
Abstract
Heavy metals can cause damage to biomolecules such as proteins and DNA in multiple ways. Cells therefore strive for keeping intracellular (heavy) metal ions bound to specific proteins that are capable of handling detoxification, export or integration as cofactors. Metal binding proteins usually provide specific coordination sites that bind certain ions with ultrahigh affinity, with the thermodynamic driving force being the stability of organometallic complexes. However, the metal binding properties of these proteins can be highly variable. Therefore the transfer of specific ions between separate proteins or even between distinct binding sites located on one and the same protein does not always follow affinity gradients, but depends on particular protein interactions that are difficult to predict. We established a method suitable to probe metal transfer between two proteins, provided the proteins are amenable to purification and in vitro handling. It consists of the loading with metals, the co-incubation and the separation of metal-exchanging proteins with subsequent determination of bound metal content. The method is exemplified by experimental data of ours probing the transfer of copper(I) between the membrane-extrinsic metal binding domain MBD2 and the transmembrane domain of CopA, a copper export ATPase from Escherichia coli (Drees et al., 2015).
Keywords: Copper-binding proteinMaterials and Reagents
- Desalting NAP5 columns (e.g., GE Healthcare, HiTrap Desalt, catalog number: 17-0853-01 )
- Gel filtration columns
We used prepacked GE 10/300 Tricorn columns, a Superdex 75 (GE Healthcare, catalog number: 17-5174-01 ) for small soluble proteins, or a Superose 6 for lipid-reconstituted samples (GE Healthcare, catalog number: 17-5172-01 ). - Proteins
Notes:- Metal binding proteins of interest must be on the hand in purified native state, with aggregates having been removed prior to the experiments.
- Proteins should be available in milligram quantities and should preferably have different molecular weights (which is a prerequisite for gel filtration).
- Proteins too small to be distinguished from each other on gel filtration columns could be fused to a polypeptide tag, e.g., the maltose binding protein or glutathione-S-transferase in order to artificially increase the molecular weight difference of the metal ion transfer pair.
- We used an eGFP tag because it is relatively inert in redox-reactions and facilitates easy detection and concentration determination.
- Care has to be taken with the use of a His-tag for means of protein purification. Although it was demonstrated that His-tags do not interfere with copper binding and quantification experiments under certain conditions (González-Guerrero et al., 2008), the metal chelating property of the hexahistidine sequence may cause metal carry-overs in some experiments.
- Metal binding proteins of interest must be on the hand in purified native state, with aggregates having been removed prior to the experiments.
- 3-(N-morpholino) propanesulfonic acid (MOPS), buffer grade (Sigma-Aldrich, catalog number: M1254 )
- Sodium chloride (Sigma-Aldrich, catalog number: S7653 )
- Sodium ascorbate (Sigma-Aldrich, catalog number: 11140 )
- Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: 43815 )
- Phosphatidylcholine (PC) (Sigma-Aldrich, catalog number: P3644 )
- Sodium dodecylsulfate (SDS) (Sigma-Aldrich, catalog number: L6026 )
- Bathocuproinedisulfonate (BCS) (Sigma-Aldrich, catalog number: B1125 )
- Bicinchoninic acid (BCA) (Sigma-Aldrich, catalog number: D8284 )
- β-dodecylmaltoside (β-DDM) (Sigma-Aldrich, catalog number: D5172 )
- Copper sulfate (CuSO4) (Sigma-Aldrich, catalog number: 451657 )
- Buffer A (see Recipes)
- BCA-copper buffer (see Recipes)
- SDS solution (see Recipes)
- Ascorbate solution (see Recipes)
- BCS solution (see Recipes)
Equipment
- (Optional) A dynamic light scattering instrument [e.g., High performance particle sizer (Malvern Instruments, model: HPP 5001 or Zetasizer Nano S )]
- Ultrasonic homogenizer for preparation of lipid emulsions (e.g., BANDELIN electronic GmbH & Co, model: UP 400 S )
- Ultrasonic bath for buffer degassing (e.g., BANDELIN electronic GmbH & Co, model: Sonorex DL510H )
- An automated chromatography unit (e.g., GE Healthcare, Äkta Purifier or Explorer)
Note: It is essential for the experiments. Use of a regular HPLC unit with fraction collector would be just as fine, provided that some modifications with respect to the tolerance of salt-containing elution buffers have been taken care of. - A thermostated shaking incubator for 1.5 ml type reaction vials (e.g., Eppendorf, catalog number: 5384000012 )
- Centrifugal evaporator (e.g., Eppendorf, catalog number: 5305000304 )
- Calibrated micropipette (e.g., Eppendorf, catalog number: 3120000062 ), alternatively: analytical balance (e.g., Mettler-Toledo International Inc., catalog number: 11142056 )
- UV-VIS Spectrophotometer (e.g., Jasco, model: J-650 ) and Nanodrop spectrophotometer (Thermo Fisher Scientific), alternatively: UV-VIS spectrophotometer and small volume cuvette (e.g., Hellma, model: 105.250-QS or Eppendorf, µCuvette®, model: G1.0 )
- Quartz microcuvette (e.g., Hellma, catalog number: 104-10-40 104-QS )
- Standard SDS-PAGE equipment (e.g., Bio-Rad Laboratories, catalog number: 1658001FC )
Procedure
- Required amount of protein
- The required amount of protein depends on how much metal donor and acceptor protein bind relative to their molecular weight.
- Using 100 nM of each donor and acceptor protein (1:1 stoichiometry) is a solid starting point.
- Lower amounts can be used, but accuracy will decrease.
- The required amount of protein depends on how much metal donor and acceptor protein bind relative to their molecular weight.
- Protein oxidation state
- Check the putative protein metal binding sites of donor and acceptor proteins for oxidation sensitivity: Monovalent copper (amongst other heavy metal ion species) is often coordinated via cysteine thiols, which is prone to oxidation during protein preparation. In our example, the donor protein, MBD2, as well as the acceptor protein CopA feature copper binding sites involving oxidation-sensitive cysteines (Fan and Rosen, 2002; Fan et al., 2001).
- In case of oxidation sensitivity of either protein, add DTT from a 1 M stock solution (one may alternatively try 2-mercaptoethanol or TCEP, but we have not tested these) to a concentration of 5-20 mM and incubate for 1 h.
- Subsequently remove all the DTT by buffer exchange via desalting chromatography using NAP5 columns (alternatively, HiTrap desalt or PD10 columns will work as well). The handling should be performed according to manufacturer’s instructions.
- In addition, it is crucial to use freshly degassed buffer (e.g., use the storage buffer established for your protein of interest, or buffer A) to avoid reoxidation.
- After desalting, proceed immediately with your experiment, or flash-freeze the pre-treated protein in liquid nitrogen.
- Check the putative protein metal binding sites of donor and acceptor proteins for oxidation sensitivity: Monovalent copper (amongst other heavy metal ion species) is often coordinated via cysteine thiols, which is prone to oxidation during protein preparation. In our example, the donor protein, MBD2, as well as the acceptor protein CopA feature copper binding sites involving oxidation-sensitive cysteines (Fan and Rosen, 2002; Fan et al., 2001).
- Load the donor protein with metal (workflow illustrated in Figure 1)
- Copper in its divalent state (such as in CuSO4) is detrimental for proteins as it adsorbs to biosurfaces and oxidizes amide bonds. Hence, protein precipitation is often observed in the presence of Cu(II) salts.
- On the contrary, Cu(I) salts are highly redox-active and are almost insoluble in aqueous media. It is therefore recommended to load the donor protein by means of a colored low affinity organo-copper complex, such as [BCA2-Cu(I)]3-. The donor protein is therefore titrated with BCA-copper buffer until the mixture does not destain any longer and retains the characteristic violet color.
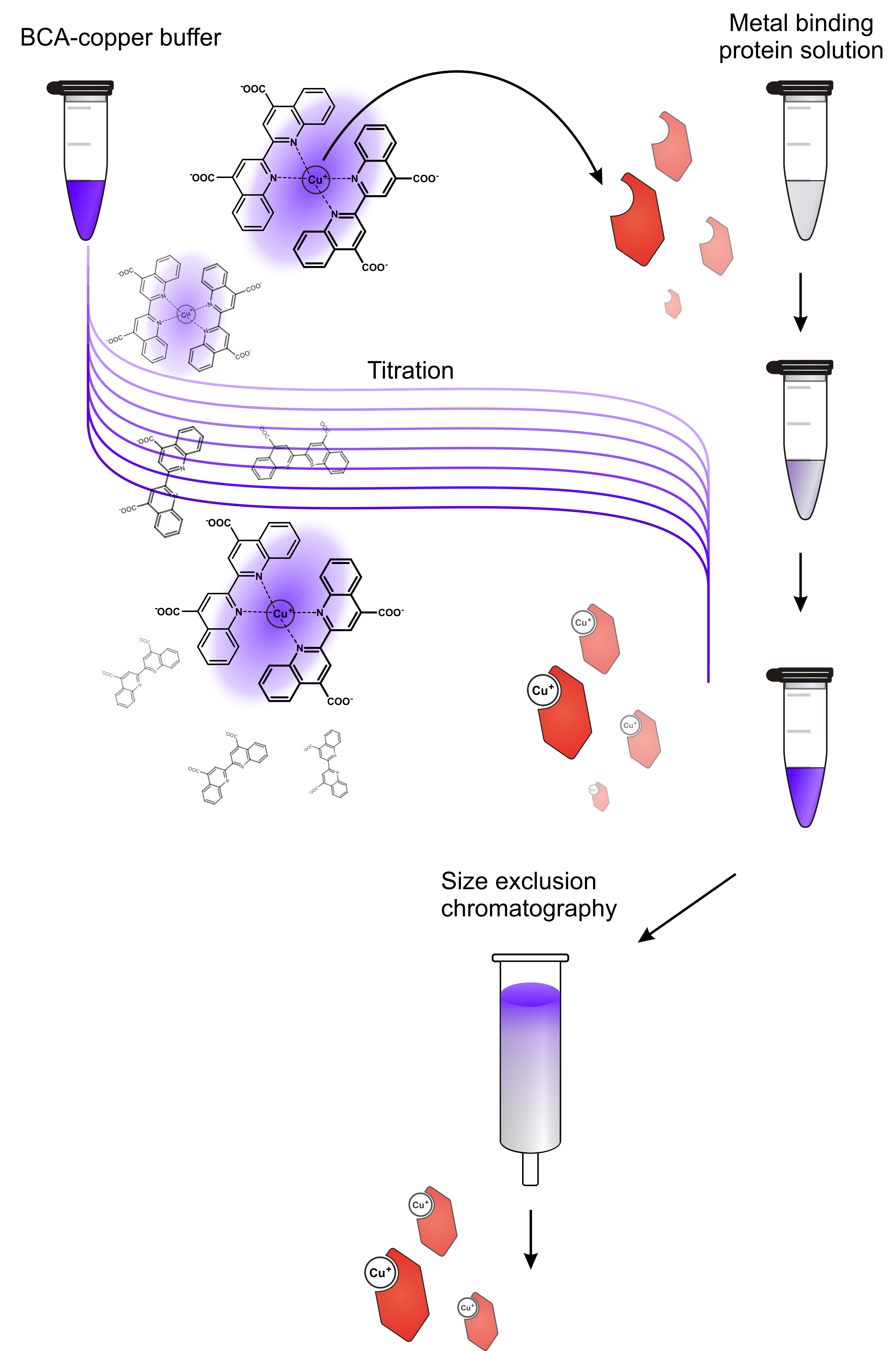
Figure 1. Workflow for preparation of metal-loaded donor protein. A solution of the donor protein in apo state is titrated with a metal buffer, in this case a BCA-copper buffer. This indirect loading procedure avoids direct contact of the protein with excess copper and thereby protects it from metal-caused damage. Titrating little steps is advantageous because it allows monitoring the loading process. In case there is no destaining in the first titration steps, the donor protein may have suffered damage or is not receptive for metal loading at all. As soon as an excess of metal buffer is present in the protein solution, the mixture is loaded onto a size exclusion/desalting column (e.g., a NAP-5 column, used after manufacturer’s instructions) to yield the metal-loaded donor protein without contamination of free metal or metal chelator. - Copper in its divalent state (such as in CuSO4) is detrimental for proteins as it adsorbs to biosurfaces and oxidizes amide bonds. Hence, protein precipitation is often observed in the presence of Cu(II) salts.
- Repurify donor protein
- To remove excess BCA, repurify the donor protein using a buffer exchange column (as in step 2c) equilibrated with buffer A.
- The eluted protein must not contain any blue stain (measure absorbance at 562 nm).
- To remove excess BCA, repurify the donor protein using a buffer exchange column (as in step 2c) equilibrated with buffer A.
- Spectroscopic determination of protein concentration
- Measure the concentration of the loaded donor protein using a NanoDrop spectrophotometer and an experimental or predicted absorption coefficient (Pace et al., 1995).
- In case the concentration of the acceptor protein is unknown, it should be quantified likewise at this point.
- Measure the concentration of the loaded donor protein using a NanoDrop spectrophotometer and an experimental or predicted absorption coefficient (Pace et al., 1995).
- (Optional) Lipid emulsion preparation
- In case of CopA, we experienced that reconstitution into lipid phase may be required for metal receptivity. Other membrane proteins (González-Guerrero et al., 2009) or soluble proteins may not require this step.
- Optimum conditions for reconstitution may vary and require testing for each individual case.
- For the presented procedure, a uniform vesicle size is advisable. Therefore, 10 % (w/v) lipid suspensions of phosphatidylcholine were emulsified by sonication for 10 min (ultrasonic homogenizer, low intensity dipped probe). The particle size distribution may be facultatively checked by means of a dynamic light scattering instrument.
- In case of CopA, we experienced that reconstitution into lipid phase may be required for metal receptivity. Other membrane proteins (González-Guerrero et al., 2009) or soluble proteins may not require this step.
- (Optional) Lipid reconstitution
Add a 10 fold (w/w) excess of the 10 % lipid suspension to a defined amount of protein and incubate at 37 °C for 20 min and gentle agitation. - Equilibration of the column
Equilibrate gel filtration column with 2 column volumes of buffer A. Meanwhile store donor and acceptor protein on ice. - Ion transfer
Mix donor and acceptor protein in molar ratio of 1:1 to 2:1 and incubate for 5 min at room temperature, then add buffer A to a total of 500 µl (for GE Healthcare Tricorn 10/300 columns). - Chromatography
Apply protein mixture to the chromatography column using the autoinjector of the unit (500 µl in 1 ml sample loop) and start the elution using a flow rate of 0.2 ml min-1 and a fraction size of 1 ml over 30 ml. An increased flow rate will work fine as well, although leading to higher dilution of the proteins. - Save the chromatogram UV and (if available) conductivity traces; it should look like the diagram in Figure 2A. Collect fractions exhibiting UV absorption and store for the later analysis of copper content (see below).
- Electrophoretic control of separation
- 10-50 µl samples can be drawn from each fraction for electrophoretic analysis of separation (perform electrophoresis according to manufacturer’s instructions).
- If donor and acceptor proteins form a stable complex during chromatography, incomplete dissociation should be observed.
- In case of transient interaction during transfer, both proteins are well separated (see Figure 2B).

Figure 2. Exemplary Data on gel filtration and electrophoresis. A. Typical UV absorption (black trace) and conductivity (gray trace) data read-out of the automated chromatography unit. The example shows a separation of MBD2 (donor) and lipid-reconstituted CopA (acceptor) using a Superose 6 column. Relevant peaks are labeled with respect to their content. B. SDS-PAGE for testing effective separation between donor and acceptor proteins is an important control experiment to rule out complex formation between donor (B-fractions) and acceptor (A-fractions). - 10-50 µl samples can be drawn from each fraction for electrophoretic analysis of separation (perform electrophoresis according to manufacturer’s instructions).
- Transfer fractions to 1.5 ml polypropylene vials and add 40 µl of SDS-solution [1% SDS (w/v) final concentration] to each fraction. Incubate at 65 °C and slight agitation until protein or lipid-protein emulsions are completely homogenized and the solutions appear clear. This can take up to 1 h.
- Cool down the samples and centrifuge for 1 min at 1,000 x g to collect the entire liquid at the bottom of the tube.
- Record UV-spectra of each sample.
- Check for the absence of a non-desired scattering curve underneath the protein absorbance spectrum.
- In case of scattering, add another 40 µl of SDS solution, incubate for 1 h and re-measure.
- Check for the absence of a non-desired scattering curve underneath the protein absorbance spectrum.
- (Optional) In case the expected metal concentrations are low, samples can be concentrated using a centrifugal evaporator (“Speed Vac”) to increase sensitivity. We experienced that concentrating to 300 µl does not cause any precipitation, but the actual level to which a sample volume can be reduced really depends on the particular experiment.
- Volume determination
Measure the exact volume of each sample, either volumetrically by using a calibrated micropipette or gravimetrically with an analytical balance [assumed density 1.02 (500 mM NaCl solution), error due to density mismatch and the precision of the pipette are in the same range]. - Detection of metal content
- Note that many colorimetric quantification methods for different metal ions are described, some of which are mentioned in the review by Xiao and Wedd (2010).
- For copper, we chose to use BCS because it has a high extinction coefficient in the visible light range. Add 1% ascorbate (v/v) and 5% BCS solution (v/v) to each sample and incubate 5 min at room temperature.
- Note that many colorimetric quantification methods for different metal ions are described, some of which are mentioned in the review by Xiao and Wedd (2010).
- Measure the absorbance of each sample at 482 nm.
- Data analysis
Calculate protein concentrations using either the computed (Pace et al., 1995) or experimental extinction coefficients. - Calculate the exact metal concentration using the extinction coefficient of the [BCS2-Cu(I)]3--complex of 13,000 M-1 cm-1 (Xiao et al., 2011). Take into account all dilution steps.
- Sum up total molar protein and copper concentrations in donor and acceptor fractions and calculate their ratios to determine the ion transfer efficiency.
- Note that in case of a reconstituted membrane protein, a 100% efficiency is very unlikely and has never been reached in our experiments. This is due to anisotropic integration of the CopA acceptor protein into the lipid phase and the resulting inaccessibility to the metal donor protein.
- With soluble proteins, transfer efficiencies of over 90% can be observed.
- Note that in case of a reconstituted membrane protein, a 100% efficiency is very unlikely and has never been reached in our experiments. This is due to anisotropic integration of the CopA acceptor protein into the lipid phase and the resulting inaccessibility to the metal donor protein.
Representative data
A typical example of the separation of copper-loaded MBD2 and of lipid-reconstituted CopA is shown in Figure 2. Full exemplary experimental data, i.e., the concentrations of protein (calculated from spectrophotometric read of OD280) and of copper (determined colorimetrically as outlined above) after the transfer experiment are displayed in Figure 4 of Reference 1.
Notes
- Avoid loading GE Superdex analytical gel filtration columns with turbid samples. Remove precipitates by centrifugation (Eppendorf microfuge) prior to sample application.
- The determinations of protein and of copper are fairly reliable. However, using analytical gel filtration columns, the sample volumes after fractionation are limiting to be sufficient for single chemical determinations of both components. Data from a single transfer experiment often provide a reasonable figure of the metal transfer efficiency. For proper validation of the transfer results, experimental data from at least four chromatographic separations should be appropriate.
- Monitoring metal contents of both, donor and acceptor proteins before the actual exchange is a very important control experiment. We therefore recommend performing gel filtrations with either donor or acceptor protein alone. Additionally, metal carryover in the receptor protein, e.g., from purification can be detected this way. The measured values can be used as a “metal content baseline” to calculate the net amount of transferred metal resulting from co-incubation.
Recipes
- Buffer A
30 mM MOPS, pH 7.5
150 mM NaCl - BCA-copper buffer
Buffer A with 2.5 mM BCA
1 mM CuSO4
20 mM ascorbate - SDS solution
25 % SDS (w/v) in H2O
Prepare at 37 °C for better solubility - Ascorbate solution
100 mM sodium ascorbate, pH adjusted to 7.5
Store in small aliquots at -20 °C - BCS solution
10 mM BCS in 30 mM MOPS, pH 7.5
Protect from light
Acknowledgments
We are grateful to the Protein Research Department of the Ruhr-Universität Bochum for financial support.
References
- Drees, S. L., Beyer, D. F., Lenders-Lomscher, C. and Lübben, M. (2015). Distinct functions of serial metal-binding domains in the Escherichia coli P1 B -ATPase CopA. Mol Microbiol 97: 423-438.
- Fan, B., Grass, G., Rensing, C. and Rosen, B. P. (2001). Escherichia coli CopA N-terminal Cys(X) (2)Cys motifs are not required for copper resistance or transport. Biochemical and Biophysical Research Communications 286: 414-418.
- Fan, B. and Rosen, B. P. (2002). Biochemical characterization of CopA, the Escherichia coli Cu (I)-translocating P-type ATPase. Journal of Biological Chemistry 277: 46987-46992.
- Gonzalez-Guerrero, M., Hong, D. and Argüello, J. M. (2009). Chaperone-mediated Cu+ delivery to Cu+ transport ATPases requirement of nucleotide binding. Journal of Biological Chemistry 284: 20804-20811.
- Gonzalez-Guerrero, M., Eren, E., Rawat, S., Stemmler, T. L. and Argüello, J. M. (2008). Structure of the two transmembrane Cu+ transport sites of the Cu+-ATPases. Journal of Biological Chemistry 283: 29753-29759.
- Pace, C. N., Vajdos, F., Fee, L., Grimsley, G. and Gray, T. (1995). How to measure and predict the molar absorption-coefficient of a protein. Protein Science 4: 2411-2423.
- Xiao, Z. G., Brose, J., Schimo, S., Ackland, S. M., La Fontaine, S. and Wedd, A. G. (2011). Unification of the copper (I) binding affinities of the metallo-chaperones Atx1, Atox1, and related proteins detection probes and affinity standards. Journal of Biological Chemistry 286: 11047-11055.
- Xiao, Z. G. and Wedd, A. G. (2010). The challenges of determining metal-protein affinities. Natural Product Reports 27: 768-789.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Drees, S. L. and Lübben, M. (2016). Analytical Gel Filtration for Probing Heavy Metal Transfer between Proteins. Bio-protocol 6(15): e1888. DOI: 10.21769/BioProtoc.1888.
Category
Biochemistry > Protein > Interaction > Protein-ligand interaction
Microbiology > Microbial biochemistry > Protein > Interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










