- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Investigating the Assembly Status of the Plastid Encoded Polymerase Using BN-PAGE and Sucrose Gradient Centrifugation
Published: Vol 6, Iss 14, Jul 20, 2016 DOI: 10.21769/BioProtoc.1873 Views: 9904
Reviewed by: Marisa RosaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Semi-throughput Procedure for Assaying Plant NADP-malate Dehydrogenase Activity Using a Plate Reader
Kevin Baudry and Emmanuelle Issakidis-Bourguet
Aug 20, 2023 1465 Views

An in vitro Assay to Probe the Formation of Biomolecular Condensates
Yu Zhang and Shen Lisha
Sep 5, 2023 3196 Views

Immunofluorescence for Detection of TOR Kinase Activity In Situ in Photosynthetic Organisms
Ana P. Lando [...] Giselle M. A. Martínez-Noël
Dec 20, 2024 1811 Views
Abstract
The plastid encoded polymerase (PEP) represents a major transcription machinery in mature chloroplasts (Liere et al., 2011; Zhelyazkova et al., 2012). The proper assembly of this multi-subunit complex is important for plant growth and development (Pfalz and Pfannschmidt, 2013). The PEP polymerase can be purified from soluble and from membrane-bound (also named transcriptionally active chromosome, TAC) fractions. Blue Native polyacrylamide gel electrophoresis (BN-PAGE) and sucrose gradient sedimentation followed by immunoblot analyses is used to detect the status of the PEP complex assembly.
Keywords: Enrichment of the plastid encoded Polymerase (PEP)Materials and Reagents
- Material
- Disposable RNase-free pipette tips (1-200 µl and 100-1,000 µl universal tips)
- RNase-free microcentrifuge tubes (1.5 ml and 2.0 ml universal tubes)
- 15 ml conical tube
- Amicon Ultra-15 centrifugal filter units, 10KDa (Merck Millipore Corporation, catalog number: UFC901024 )
- PVDF-Membrane (Carl Roth, Roti® -Fluoro, catalog number: 2803.1 )
- Polypropylene centrifuge tubes, 14 x 95 mm (Beckman, catalog number: 331374 )
- Disposable RNase-free pipette tips (1-200 µl and 100-1,000 µl universal tips)
- Plant material
- Maize seedlings grown in soil for 7-10 days at 26-28 °C in cycles of 16 h-light/8 h-dark
- Maize seedlings grown in soil for 7-10 days at 26-28 °C in cycles of 16 h-light/8 h-dark
- Antibodies
- anti-ZmpTAC12 (custom antibody) (Biogenes)
- anti-ZmRpoA (custom antibody) (Biogenes)
- Goat anti-Rabbit IgG HRP-linked (Sigma-Aldrich, catalog number: A6154 )
- anti-ZmpTAC12 (custom antibody) (Biogenes)
- Reagent
- Liquid nitrogen
- Acrylamide, Gel 40 (29:1) (Carl Roth, Rotiphorese®, catalog number: A515.1 )
- Ammonium persulfate (APS), (NH4)2S2O8 (Sigma-Aldrich, catalog number: 248614-500G-D )
- Bis-(2-hydroxyethyl)-imino-tris-(hydroxymethyl)-methane, Bis-Tris (Carl Roth, catalog number: 9140.3 )
- β-Mercaptoethanol (Carl Roth, catalog number: 4227.1 )
- Bradford, Roti®-Nanoquant (Carl Roth, catalog number: K880.1 )
- Coomassie brilliant blue G250 (Sigma-Aldrich, catalog number: B0770-5G )
- 1, 4-dithiothreitol, DTT (Carl Roth, catalog number: 6908.1 )
- ε-aminocaproic acid (Carl Roth, catalog number: 3113.2 )
- Glycerol (Carl Roth, catalog number: 6962.3 )
- Glycine (Carl Roth, catalog number: 3187.4 )
- HEPES (Carl Roth, catalog number: 9105.3 )
- Magnesium acetate tetrahydrate, Mg(CH3COO)2·4H2O, (Carl Roth, catalog number: 0 275.2 )
- Magnesium chloride, MgCl2 (Carl Roth, catalog number: KK36.3 )
- Methanol (Carl Roth, catalog number: P717.1 )
- n-Dodecyl β-D-maltoside, β-DM (Sigma-Aldrich, catalog number: D4641-1G )
- N, N, N’, N’-Tetramethylethylenediamine (TEMED) (Carl Roth, catalog number: 2367.1 )
- N-Tris-(hydroxymethyl)-methyl-glycin, Tricin (Carl Roth, catalog number: 6977.3 )
- BlueEasy Prestained Protein Marker (Nippon Genetics, catalog number: MWP06 )
- Potassium acetate, K(CH3COO) (Carl Roth, catalog number: T874.1 )
- Potassium hydroxide (Carl Roth, catalog number: 7986.1 )
- Protease inhibitor cocktail (cOmplete)
Note: Currently, it is “Sigma-Aldrich, cOmpleteTM, catalog number: 000000011836153001 ”. - Tris-(hydroxymethyl)-aminomethan, TRIS (Carl Roth, catalog number: 4855.3 )
- Triton X-100 (Carl Roth, catalog number: 3051.2 )
- Tween-20 (Carl Roth, catalog number: 9127.1 )
- Sodium chloride, NaCl (Carl Roth, catalog number: 9265.2 )
- Sodium dodecyl sulfate, SDS (Carl Roth, catalog number: 4360.2 )
- Sodium fluorid, NaF (Carl Roth, catalog number: 4503.2 )
- D-sucrose [D(+)-saccharose] Carl Roth, catalog number: 4621.2 )
- Liquid nitrogen
- Buffers
- BN-Lysis buffer (see Recipes)
- 3x BN-Gel buffer (see Recipes)
- 4.5% Separating gel (see Recipes)
- 14% Separating gel (see Recipes)
- 4% Stacking gel (see Recipes)
- Cathode buffer blue (see Recipes)
- Cathode buffer (see Recipes)
- Anode buffer (see Recipes)
- 2x BisTris ACA (ACA, aminocaproic acid) (see Recipes)
- Sample buffer (see Recipes)
- 2% n-dodecyl-β-D-maltopyranoside (β-DM) solution (see Recipes)
- BN-Loading buffer (see Recipes)
- Transfer buffer (see Recipes)
- 1x TBST buffer (see Recipes)
- Low sucrose solution (see Recipes)
- High sucrose solution (see Recipes)
- Diluent solution (see Recipes)
- BN-Lysis buffer (see Recipes)
Equipment
- Mortar and pestle
- Sonicator (BANDELIN electronic GmbH & Co., model: Sonopuls HD 2200 )
- Microcentrifuge (Beckman, model: Avanti JXN-30 )
- Ultracentrifuge (Sorvall Discovery 90SE)
Note: This product has been discontinued by the manufacturer. - SW40 rotor (Beckman)
- Gradient mixer (BIO-RAD, model: 385 )
Note: This product has been discontinued by the manufacturer. - BioPhotometer (Eppendorf)
- Electrophoresis apparatus (Hoefer, model: SE600 )
- Semi-Dry-Blotter (VWR International, PEQLAB Biotechnologie GmbH)
Procedure
- Blue Native polyacrylamide gel electrophoresis (BN-PAGE) analysis
- Collect plant tissue (basal half of one second leaf) into a 1.5 ml microcentrifuge tube and quickly freeze in liquid nitrogen. Frozen tissues can be stored at -80 °C for 1-2 months.
Note: Since the expression of PEP subunits is higher in younger than in mature chloroplasts, proteins from young leaf sections (basal half of second leaves) were used in this study. - Grind tissue in pre-chilled mortar with a pestle to a fine powder in liquid nitrogen.
- Let all liquid nitrogen evaporate and then immediately add 200-400 µl ice-cold BN-Lysis buffer directly to the tissue in the mortar. Do not let the tissue thaw before you add the BN-Lysis buffer.
- If the lysis buffer freezes, wait until it thaws before proceeding to the next step.
- Keep sample on ice and homogenize with 10-20 strokes of the pestle.
- Transfer sample into a 1.5 ml microcentrifuge tube and determine the protein concentration.
Note: The concentration should be at least 4 µg/µl. - Divide into 50-100 μl aliquots, snap freeze in liquid nitrogen and store at -80 °C for later use.
- Prepare gradient gel (gel dimension: 18 x 16 x 0.15 cm) from 14 ml of each of the two separation solutions (4% and 14% acrylamide) with a gradient mixer. Overlay gel with stacking solution prior it gets solid. Let polymerize at room temperature, and then cool the gel down to 4 °C.
- To prepare for BN-PAGE, let the sample thaw on ice and then combine 10-25 µl sample (50-100 µg) of it with 25 µl sample buffer, 17 µl 2% β-DM solution and 4.5 µl BN-Loading buffer.
Note: The Bradford assay was used to determine protein concentration. - Incubate sample on ice for 10 min.
- Centrifuge sample for 10 min at 17,500 x g and 4 °C.
- Load supernatant into wells and run the gel overnight (16-18 h) at 50 V constant voltage at 4 °C (or after the dye front has migrated about one third).
- Replace blue cathode buffer to cathode buffer without dye.
- Continue the electrophoresis at 10 mA constant current (~600 V) until the dye runs off (4-5 h).
- Remove gel from glass plates and soak gel in precooled (10 °C) transfer buffer for approx. 30 min.
- Transfer proteins onto PVDF membrane using semi-dry blotting apparatus (for 3 h at 0.8-3 mA/cm2 at room temperature).
- Destain membrane in 100% methanol for 1-3 min and immediately rinse with water.
- Block membrane overnight with 5% casein in 1x TBST buffer at 4 °C.
- Rinse membrane briefly with water and then proceed with immunoprobing according to a standard protocol (Sambrook and Russell, 2001).
- Collect plant tissue (basal half of one second leaf) into a 1.5 ml microcentrifuge tube and quickly freeze in liquid nitrogen. Frozen tissues can be stored at -80 °C for 1-2 months.
- Sucrose gradient sedimentation analysis. The protocol for sample preparation described above can be used with the following modifications:
- Harvest tissue from 2-3 seedlings (second whole leaves).
- Use 1.5 ml buffer for lysis (step A3).
- After homogenization (step A5) on ice transfer sample into a 15 ml conical tube.
- Sonicate sample (tip width 2-3 mm) for three cycles of 10 sec at 20% power output. Let samples sit on ice for 1-2 min between sonication (Note 1).
- Transfer lysate into a 2.0 ml microcentrifuge tube and centrifuge at 20,000 x g for 30 min at 4 °C to remove insoluble material. Take an aliquot of each fraction and detect individual PEP subunits by immunoblot analysis (Note 1).
- Transfer supernatant into a fresh 15 ml tube and determine protein concentration.
Note: Concentration should be at least 3 µg/µl. - Snap freeze sample (supernatant) in liquid nitrogen and store at -80 °C for later use.
- Thaw samples on ice for > 30 min and add 1.5 ml diluent solution. Diluent solution is used to dilute the glycerol concentration in the sample prior to sucrose gradient centrifugation.
- Concentrate sample to 1.0 ml using cut-off centrifugal filter device, when necessary.
- Prepare 12-ml continuous gradients of 10 to 30% sucrose (from 6 ml each) in 14 ml polyallomer centrifugal tubes with a gradient mixer (sucrose gradient preparation in Zhu, 2012).
- Load slowly 1.0 ml sample (approx. 5 mg) on top of a gradient and centrifuge tubes for 15 h at 146,000 x g in an SW-40 rotor (4 °C).
- Carefully collect 500-µl-fractions from the top of the gradient.
- Use equal volumes (30-50 µl) of every second fraction for SDS-PAGE and subsequent immunoblot analysis with an antibody specific for a PEP subunit.
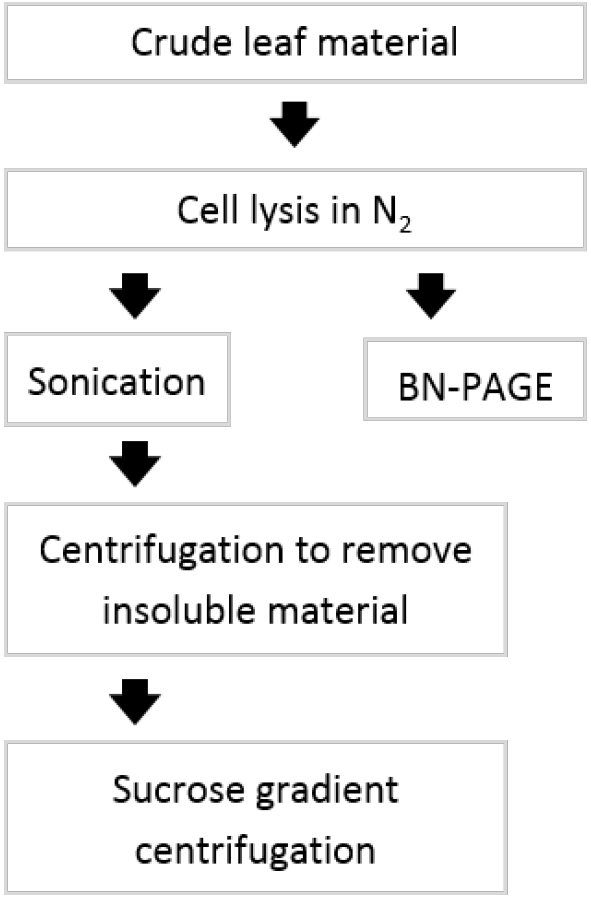
Figure 1. Flowchart of experimental procedure
- Harvest tissue from 2-3 seedlings (second whole leaves).
Representative data

Figure 2. Separation of protein complexes by BN-PAGE and subsequent analyses of PEP complex assembly by immunoblotting with antibodies directed against the subunits RpoA and pTAC12. Approximately 50 µg of total leaf protein from the base section of second leaves were loaded per lane. RpoA, RNA polymerase alpha subunit; pTAC12, plastid Transcriptionally Active Chromosome 12 (Note 2).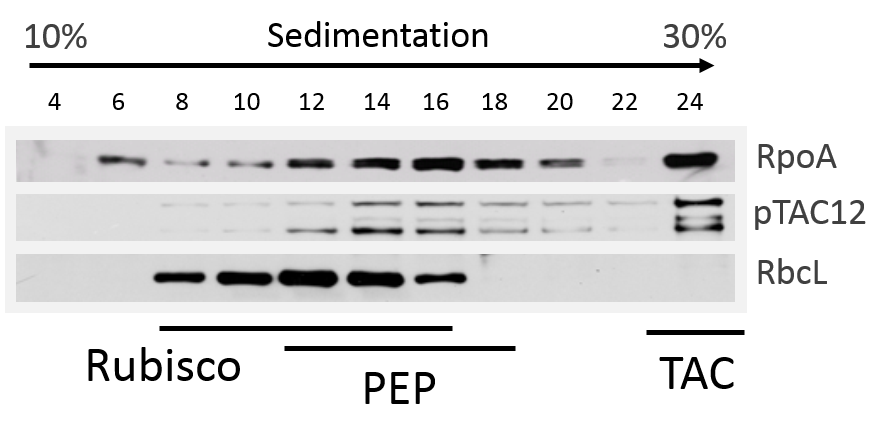
Figure 3. Sucrose-gradient sedimentation. Total leaf proteins from basal half of second leaf were solubilized in BN-Lysis buffer with sonication and soluble fractions subjected to sucrose gradient centrifugation for analysis of PEP-complex assembly (Pfalz et al., 2014). Fractions were collected and immunoblotted with antibodies against pTAC12, RpoA and Rubisco (RbcL). Respective fractions containing Rubisco, PEP (plastid-encoded RNA polymerase), and TAC (transcriptionally active chromosome) (Note 2) are indicated by bars.
Notes
- The DNA-bound PEP fraction (TAC) is tightly associated with the thylakoid membranes. It can be released by mild sonication. To validate efficient release of PEP from membranes by sonication take an aliquot of each fraction and detect individual PEP subunits by immunoblot analysis.
- The TAC complex does not enter the separation gel on conventional BN-Gels, and therefore cannot be immunodetected. The detected proteins by BN-PAGE represent the soluble PEP complex. It is expected to see a shift towards small size fractions if the complex is partially assembled. In case that the proteins are not properly assembled into the full PEP complex the appropriate bands will disappear (PEP: ~1,000 kDa).
Recipes
Note: All buffers should be stored at 4 °C prior use unless otherwise noted.
- BN-Lysis buffer
100 mM Tris-HCl (pH 7.3)
10 mM MgCl2
25% (v/v) glycerol
1% (v/v) Triton X-100
5 mM β-mercaptoethanol
1x Protease inhibitor cocktail - 3x BN-Gel buffer
150 mM Bis-Tris (pH 7.0)
1.5 M ε-aminocaproic acid - 4.5% Separating gel (prepare fresh)
1.80 ml Acrylamide/Bisacrylamide (29:1)
5.40 ml 3x BN-Gel Buffer
1.80 ml 75% Glycerol
7.13 ml dH2O
35 μl 10% APS
35 μl TEMED - 14% Separating gel (prepare fresh)
5.60 ml Acrylamide/Bisacrylamide (29:1)
5.40 ml 3x BN-Gel Buffer
4.6 ml 75% glycerol
330 µl dH2O
35 μl 10% APS
35 μl TEMED - 4% Stacking gel (prepare fresh)
2.1 ml Acrylamide/Bisacrylamide (29:1)
7.00 ml 3x BN-Gel Buffer
75% glycerol
11.79 ml dH2O
50 µl 10% APS
60 µl TEMED - Cathode buffer blue
0.5 M tricine
150 mM Bis-Tris (pH 7.0)
0.02% Coomassie blue G250 - Cathode buffer
0.5 M tricine
150 mM Bis-Tris (pH 7.0) - Anode buffer
0.5 M Bis-Tris (pH 7.0) - 2x BisTris ACA (can be stored at -20 °C)
200 mM Bis-Tris (pH 7.0)
1 M ε-aminocaproic acid - Sample buffer (can be stored at -20 °C)
125 µl 100 mM Bis-Tris (pH 7.0)
250 µl 40% glycerol
20 µl 25x Protease inhibitor cocktail
5 µl 1M NaF
100 µl dH2O - 2% n-dodecyl-β-D-maltopyranoside (β-DM) solution (can be stored at -20 °C)
125 µl 100 mM Bis-Tris (pH 7.0)
500 µl 40% glycerol
200 µl 10% β-DM (in dH2O)
50 µl 25x Protease inhibitor cocktail
125 µl dH2O - BN-Loading buffer (can be stored at -20 °C)
50 mg Coomassie blue G250
500 µl 2x BisTris ACA
400 µl 75% saccharose
50 µl dH2O - Transfer buffer
0.5% SDS
25 mM Tris-HCl (pH 7.6)
192 mM glycine
10% methanol - 1x TBST
25 mM Tris-HCl (pH 7.5)
125 mM NaCl
0.10% Tween-20 - Low sucrose solution (should be made fresh each time and stored at 4 °C prior to use)
10% (w/v) sucrose
30 mM HEPES-KOH (pH 8.0)
100 mM KOAc
10 mM Mg(OAc)2
5 mM dithiothreitol - High sucrose solution (should be made fresh each time and stored at 4 °C prior to use)
30% (w/v) sucrose
30 mM HEPES-KOH (pH 8.0)
100 mM KOAc
10 mM Mg(OAc)2
5 mM dithiothreitol - Diluent solution (can be stored at -20 °C)
30 mM HEPES-KOH (pH8.0)
200 mM KOAc
20 mM Mg(OAc)2
2 mM dithiothreitol
1x Protease inhibitor cocktail
Acknowledgments
The work was supported by the DFG (PF667-4). This work is adapted from the work previously published (Pfalz et al., 2015). BN-PAGE is performed essentially as described by Schägger and von Jagow.
References
- Liere, K., Weihe, A. and Borner, T. (2011). The transcription machineries of plant mitochondria and chloroplasts: Composition, function, and regulation. J Plant Physiol 168(12): 1345-1360.
- Pfalz, J. and Pfannschmidt, T. (2013). Essential nucleoid proteins in early chloroplast development. Trends Plant Sci. 18(4): 186-194.
- Pfalz, J., Holtzegel, U., Barkan, A., Weisheit, W., Mittag, M. and Pfannschmidt, T. (2015). ZmpTAC12 binds single-stranded nucleic acids and is essential for accumulation of the plastid-encoded polymerase complex in maize. New Phytol 206(3): 1024-1037.
- Sambrook, J. and Russell, D. W. (2001). Molecular cloning: A laboratory manual. 3rd edition. Cold Spring Harbor.
- Schagger, H. and von Jagow, G. (1991). Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem 199(2): 223-231.
- Zhelyazkova, P., Sharma, C. M., Forstner, K. U., Liere, K., Vogel, J. and Borner, T. (2012). The primary transcriptome of barley chloroplasts: numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell 24(1): 123-136.
- Zhu, H. (2012). Sucrose gradient analysis. Bio-protocol Bio101: e194.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Pfalz, J. (2016). Investigating the Assembly Status of the Plastid Encoded Polymerase Using BN-PAGE and Sucrose Gradient Centrifugation . Bio-protocol 6(14): e1873. DOI: 10.21769/BioProtoc.1873.
Category
Biochemistry > Protein > Isolation and purification
Plant Science > Plant biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









