- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Bioassay Protocol for Quorum Sensing Studies Using Vibrio campbellii
Published: Vol 6, Iss 14, Jul 20, 2016 DOI: 10.21769/BioProtoc.1866 Views: 11812
Reviewed by: Arsalan DaudiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In vivo Analysis of Cyclic di-GMP Cyclase and Phosphodiesterase Activity in Escherichia coli Using a Vc2 Riboswitch-based Assay
Ying Liu [...] Ute Römling
Mar 5, 2018 7913 Views

Extraction of Small Molecules from Fecal Samples and Testing of Their Activity on Microbial Physiology
Eduardo S. Alves [...] L. Caetano M. Antunes
Apr 20, 2018 7986 Views

Analysis of Gram-negative Bacteria Peptidoglycan by Ultra-performance Liquid Chromatography
Laura Alvarez [...] Felipe Cava
Oct 5, 2020 5449 Views
Abstract
Quorum Sensing (QS), or bacterial cell-to-cell communication, is a finely-tuned mechanism that regulates gene expression on a population density-dependent manner through the production, secretion and reception of extracellular signaling molecules termed autoinducers (AIs). Given that QS plays an important role in bacterial biofilm formation and virulence factor production in many pathogenic strains, QS disruptors have become a hot topic in current antimicrobial research. There are several reporter strains exhibiting QS-regulated phenotypes that have been engineered for the identification of QS inhibitors, including, for example, pigment production (González and Keshavan, 2006; Steindler and Venturi, 2007), gfp, lacZ or lux reporter gene fusions (González and Keshavan, 2006; Steindler and Venturi, 2007), or lethal gene fusions downstream QS-controlled promoters (Weiland-Bräuer et al., 2015). With three parallel QS circuits, the bioluminescent marine bacterium Vibrio campbellii (formerly harveyi, Lin et al., 2010) constitutes a complex Gram-negative model for which an extensive body of knowledge exists, including an array of mutant biosensors. In V. campbellii, bioluminescence is regulated by QS. However, bioluminescence is the result of complex biochemical networks that converge with cell respiration and fatty acid metabolism. It is also an energy-demanding reaction that strongly depends on the overall metabolic state of the bacterium, consuming up to 1/5 of the cell resources (Munn, 2011). Thus, disruption of QS-controlled phenotypes might be the result of toxic side effects or interference with the above-mentioned biochemical pathways rather than QS signaling. Therefore, adequate control experiments should be included. The protocol described herein provides a method and workflow for the identification of putative QS-disrupting compounds in Vibrio. It can also be easily adapted for other QS studies (e.g., detection of AI molecules).
Keywords: Quorum SensingMaterials and Reagents
- White, clear-bottom 96-well plates (Sigma-Aldrich, Corning® Costar®, catalog number: 3610 )
- Sterile sealing membrane (Sigma-Aldrich, Breathe-Easy®, catalog number: Z380059 )
- 15-ml tubes (VWR international, catalog number: 525-0150 )
- 96-well plate (Thermo Fisher Scientific, NuncTM MicroWellTM, catalog number: 167008 )
- Cryovials (optional) (Cryoinstant) (Deltalab, catalog number: 409113/6 )
- Disposable 1 mm path length cuvettes (Sigma-Aldrich, Brand®, catalog number: Z637092 )
- Vibrio campbellii strain
- Dimethylsulfoxide (DMSO) (Sigma-Aldrich, catalog number: D8418 )
- NaCl (Sigma-Aldrich, catalog number: S7653 )
- MgSO4 (Sigma-Aldrich, catalog number: M7506 )
- Protein Hydrolysate Amicase (Sigma-Aldrich, catalog number: 82514 )
- DI water
- NaOH (Sigma-Aldrich, catalog number: S5881 )
- Glycerol
- Potassium phosphate monobasic (Sigma-Aldrich, catalog number: P5655 )
- Potassium phosphate dibasic (Sigma-Aldrich, catalog number: 1551128 )
- L-arginine (Sigma-Aldrich, catalog number: A5006 )
- Kanamycin (Sigma-Aldrich, catalog number: K1377 )
- Cinnamaldehyde (Sigma-Aldrich, catalog number: W228613 )
- Marine Agar (MA) (Conda, catalog number: 1217 )
- Autoinducer Bioassay (AB) medium (see Recipes)
Equipment
- EnSpire® Multimode Plate Reader (PerkinElmer, model: 2300-0000 )
- Tube rotator (Grant-bio, model: PTR35 )
- Spectrophotometer (Beckman Coulter, model: DU 530 )
Software
- GraphPad Prism (GraphPad Software, Inc.)
- Microsoft Excel (Microsoft Corporation)
Procedure
- Streak a MA plate with the cryopreserved V. campbellii strain of interest. Incubate for single colonies at 30 °C for 24-36 h. If required, supplement the plates with suitable antibiotics (e.g., kanamycin, see flow chart in Figure 4 for more details).
Note: We generally prepare two bacterial stocks: the first in commercial cryovials with beads for storage in our strain repository, and the second in 25-30% glycerol to be used as a working stock. Both are preserved at -80 °C. Even if we have not experienced any issue, we use this copy of the stock since repeated freezing and thawing can damage the cells, particularly if they are used routinely.
For one streak we just use a 1-μl inoculating loop or any other sterile material. We start the assays from single colonies after 24-36 h incubation. This agar plate can be preserved at 4-5 °C for at least 1 week without loss of reproducibility. Alternatively, cultures can be prepared directly from the stocks; in that case, we recommend using 10 μl for inoculating 3 ml of AB medium (see next step). - Prepare a culture of the V. campbellii strain of interest by inoculating 3 ml of AB medium in a 15-ml tube with a fresh colony. Incubate overnight in a tube rotator (40 rpm) at 30 °C to an OD600 of around 0.5 (1 mm path length).
- Distribute 100 µl of a 2x dilution of the test compounds in AB medium into the wells of a white, clear-bottom 96-well plate. Serial dilutions, if required, can be prepared in the plate.
Note: We usually prepare 80 mM stocks of the test products in DMSO. Using a cut-off dose of 200 µM yields a maximum solvent concentration of 0.25% v/v that has not significant effect on bacterial growth and luminescence.
No-treatment controls and solvent controls are included in each plate, as well as 100 μM cinnamaldehyde (positive control) and water (negative control) (Figure 1).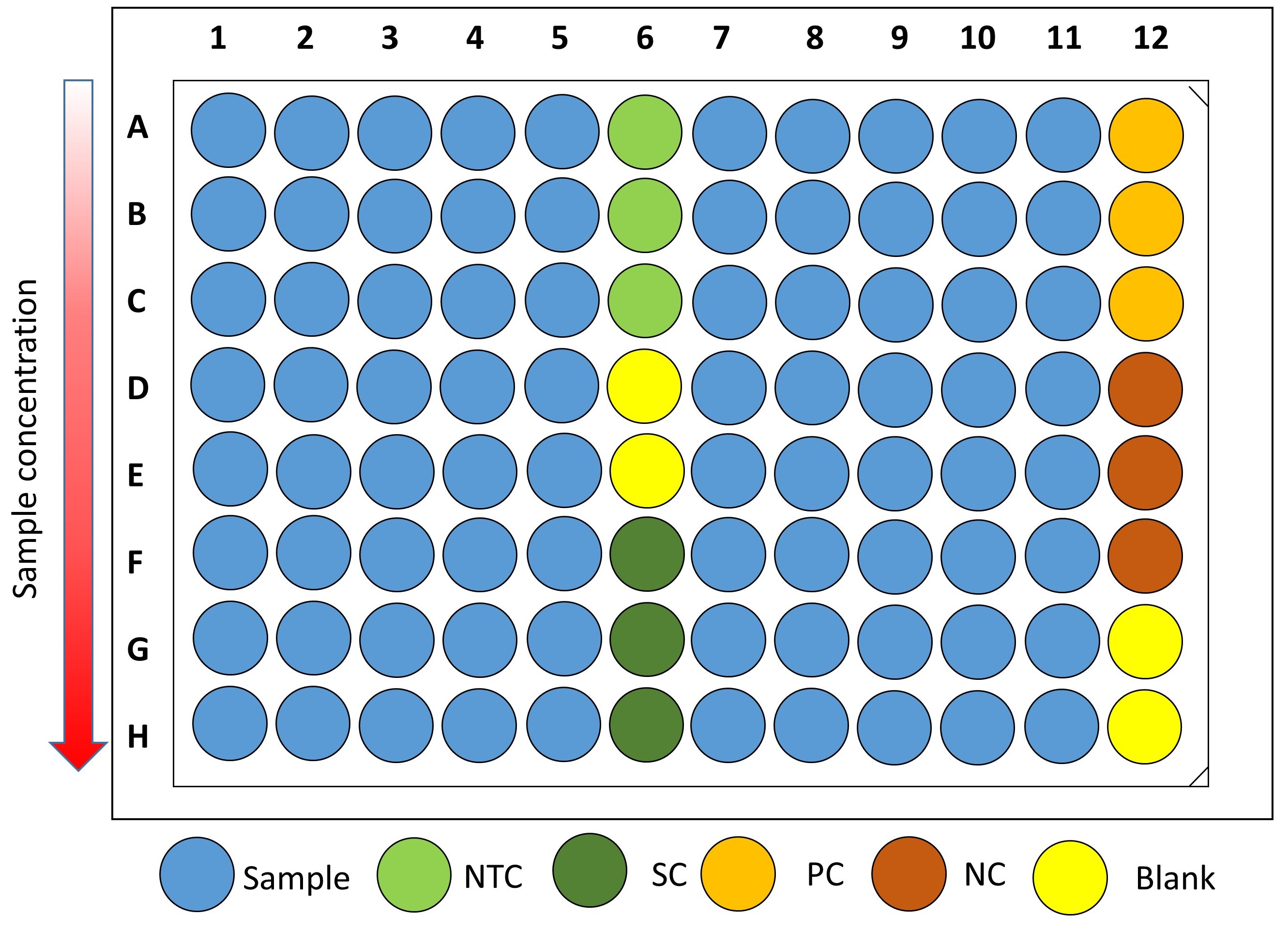
Figure 1. Plate distribution showing sample wells (x80), No-Treatment Control wells (NTC, x3), Solvent Control wells (SC, x3), Positive Control wells (PC, x3), Negative Control wells (NC, x3) and Blank wells (x4). Serial two-fold dilutions of the test compounds, if required, are proposed from rows A to H. - Dilute the overnight V. campbellii culture 1:50 in fresh AB medium. This gives an inoculum of approximately 1-2 x 107 CFU ml-1 that will be further diluted 1:2 (see next step).
Note: When establishing this protocol in the laboratory, we recommend verifying the cell density of the inoculum as described by Harrison et al., 2010. Briefly, 180 µl of AB medium are distributed into four columns of a sterile 96-well plate, from row A to H. Twenty µl of inoculum are pipetted into the A-row wells and serially diluted (-1 to -8 dilutions). Using a multichannel pipette, 10 µl of the dilutions are spotted onto the surface of a MA plate. Up to four replicates of each dilution can be easily spotted onto a standard 90-100 mm agar plate. The spots are dried inside a biosafety cabinet and the plates incubated for 24 h at 30 °C for CFU count. For an accurate determination of the cell density, take into consideration the lowest concentration in which individual colonies can be counted. The CFU per ml will be =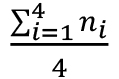 x 10 (d+2), where n represents the colony count in each of the 4 replicates, and d the corresponding dilution. The actual cell density in the assay will be half of this calculation.
x 10 (d+2), where n represents the colony count in each of the 4 replicates, and d the corresponding dilution. The actual cell density in the assay will be half of this calculation. - Distribute 100 µl of the diluted bacterial culture into the wells.
- Seal the plate with a sterile, breathable and transparent sealing film (Figure 2).
Note: Use a brayer for carefully sealing the plate. Do not remove the external protective cover until the plate is properly sealed in order to avoid contamination.
Figure 2. Aspect of a sealed, white, clear-bottom 96-well plate
- Incubate the plate for 18 h at 30 °C and internal orbital shaking (150 rpm). Take OD600 and luminescence readings every 15 min.
Note: Plate dimension, measurement height and Z’ optimizations are advised when using EnSpire® Multimode Plate Reader. Similarly, flatfield and crosstalk corrections optimize the readings. We do not recommend using a distance between plate and detector lower than 0.6 mm (otherwise, contaminations might occur due to damage of the sealing film). Activate the condensation prevention option for sealed plates (upper heater temperature 2 °C warmer than lower heater temperature).
Data analysis
- Subtract the background values for OD600 and luminescence (Microsoft Excel)
Note: In luminescence readings, cross-talk differs slightly in function of the position of the wells. Thus, central wells are more susceptible to well-to-well light cross-talk. A tentative plate distribution that takes into account this effect is shown in Figure 1. Optimization of measurement parameters diminishes this phenomenon, although it takes place even with proper optimization. However, luminescence readings in blank wells are always at least two orders of magnitude lower than those recorded for sample wells and consequently, cross-talking exerts a negligible effect with our experimental settings. - Using a scientific graphing software (e.g., GraphPad Prism), represent the time-course curves for bacterial growth and bioluminescence (Figure 3). Determine the area below the curves and calculate the relative inhibition of the treatments with respect to the controls for each variable.
Note: It could be useful to normalize the luminescence data to the OD600 for each dose or treatment, i.e., NL=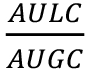 where NL = Normalized Luminescence, AULC = Area Under Luminescence Curve and AUGC = Area Under Growth Curve.
where NL = Normalized Luminescence, AULC = Area Under Luminescence Curve and AUGC = Area Under Growth Curve.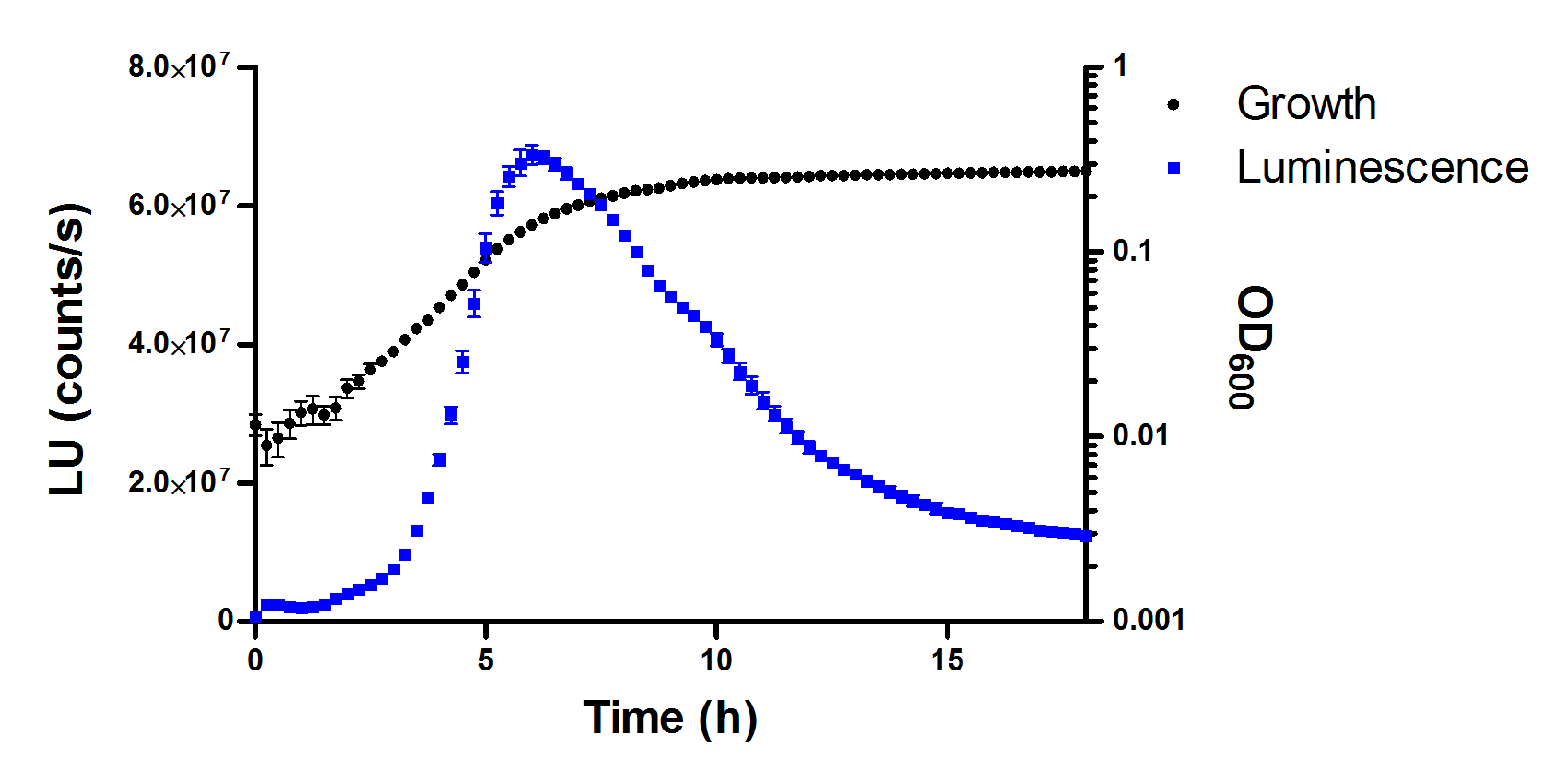
Figure 3. Chart displaying a typical NTC growth curve (black squares) and, overlapped, the corresponding luminescence curve (blue squares) over an assay period of 18 h. Data represent the mean and standard deviation of three replicates. - With these data, calculate the corresponding IC50 values using non-linear regression (GraphPad Prism).
- Proposed workflow for the discovery of Quorum Sensing Inhibitors (Figure 4).
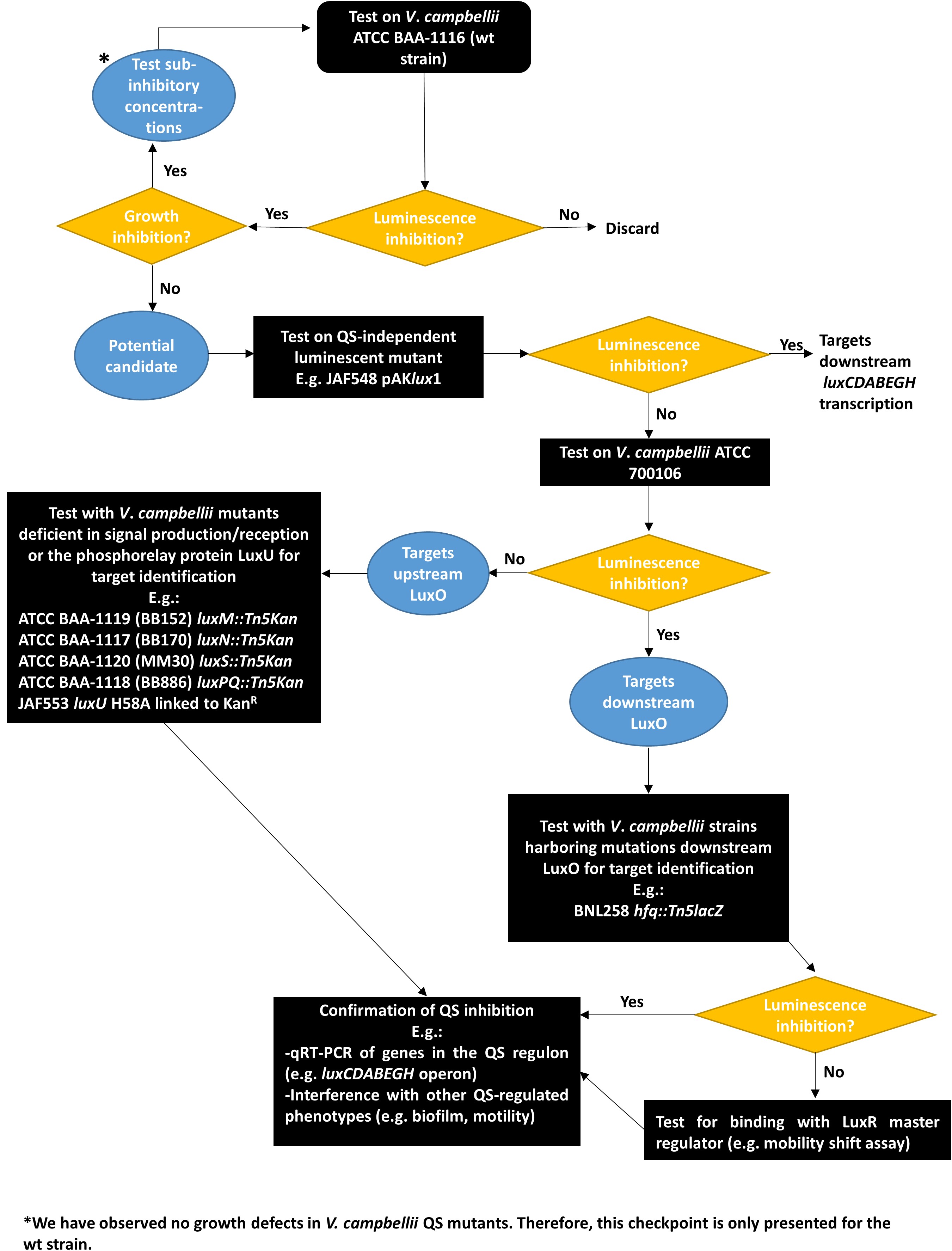
Figure 4. Flowchart for testing and characterizing the activity of putative Quorum Sensing Inhibitors using Vibrio campbellii QS mutants
Recipes
- AB Medium
17.5 g NaCl
12.3 g MgSO4
2.0 g casamino acids
970 ml DI water
Dissolve the ingredients and bring the pH of the solution to 7.5 with 3 N NaOH
Autoclave at 121 °C and after completely cooling down, add the following ingredients from separate filter-sterilized stocks:
10 ml 1 M potassium phosphate (pH 7.0)
10 ml 0.1 M L-arginine
10 ml glycerol
Acknowledgments
This protocol was developed with financial support provided by the Spanish Ministry of Economy and Competitiveness, grant CTQ2014-55888-C03-01. A.J.M-R. holds a fellowship from PLOCAN. This protocol is an adaptation from that published in Martín-Rodríguez et al. 2015a and 2015b.
References
- González, J. E., Keshavan, N. D. (2006). Messing with bacterial Quorum Sensing. Microbiol Mol Biol Rev 70(4): 859-875.
- Harrison, J. J., Stremick, C. A., Turner, R. J., Allan, N. D., Olson, M. E. and Ceri, H. (2010). Microtiter susceptibility testing of microbes growing on peg lids: a miniaturized biofilm model for high-throughput screening. Nat Protoc 5(7): 1236-1254.
- Lin, B., Wang, Z., Malanoski, A. P., O'Grady, E. A., Wimpee, C. F., Vuddhakul, V., Alves Jr, N., Thompson, F. L., Gomez-Gil, B. and Vora, G. J. (2010). Comparative genomic analyses identify the Vibrio harveyi genome sequenced strains BAA-1116 and HY01 as Vibrio campbellii. Environ Microbiol Rep 2(1): 81-89.
- Munn, C. (2011). Marine Microbiology. 2nd ed. New York: Garland Science: 105.
- Martín-Rodríguez A. J., Babarro J. M. F., Lahoz F., Sansón M., Martín V. S., Norte M., Fernández J. J. (2015). From broad-spectrum biocides to Quorum Sensing disruptors and mussel repellents: antifouling profile of alkyl triphenylphosphonium salts. PLoS One 10(4): e0123652.
- Martín-Rodríguez A. J., Ticona J. C., Jiménez I. A., Flores N., Fernández J. J., Bazzocchi I. (2015). Flavonoids from Piper delineatum modulate Quorum-Sensing regulated phenotypes in Vibrio harveyi. Phytochemistry 117: 98-106.
- Steindler, L., Venturi, V. (2007). Detection of Quorum-Sensing N-acyl homoserine lactone signal molecules by bacterial biosensors. FEMS Microbiol Lett 266(1): 1-9.
- Weiland-Bräuer N., Pinnow, N., Schmitz R. A. (2015). Novel reporter for identification of interference with acyl homoserine lactone and autoinducer-2 Quorum Sensing. J Bacteriol 81(4): 1477-1489.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Martín-Rodríguez, A. J. and Fernández, J. J. (2016). A Bioassay Protocol for Quorum Sensing Studies Using Vibrio campbellii. Bio-protocol 6(14): e1866. DOI: 10.21769/BioProtoc.1866.
Category
Microbiology > Microbial signaling > Quorum sensing
Microbiology > Microbial cell biology > Cell-based analysis
Biochemistry > Other compound > Flavonoid
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











