- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
MNase Digestion for Nucleosome Mapping in Neurospora
Published: Vol 6, Iss 11, Jun 5, 2016 DOI: 10.21769/BioProtoc.1825 Views: 10305
Reviewed by: Valentine V TrotterEmily CopeEmilia Krypotou

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Measuring UV-induced Mutagenesis at the CAN1 Locus in Saccharomyces cerevisiae
Ildiko Unk and Andreea Daraba
Oct 20, 2014 12761 Views

Using CRISPR/Cas9 for Large Fragment Deletions in Saccharomyces cerevisiae
Huanhuan Hao [...] Liping Zhang
Jul 20, 2017 14244 Views

Method for Multiplexing CRISPR/Cas9 in Saccharomyces cerevisiae Using Artificial Target DNA Sequences
Rachael M. Giersch and Gregory C. Finnigan
Sep 20, 2017 13368 Views
Abstract
Digestion of chromatin by micrococcal nuclease MNase followed by high throughput sequencing allows us to determine the location and occupancy of nucleosomes on the genome. Here in this protocol we have described optimized conditions of MNase digestion of filamentous fungus Neurospora crassa chromatin without a requirement of a nuclear fractionation step.
Keywords: Neurospora crassaMaterials and Reagents
- Filter paper (VWR International, catalog number: 1001-090 )
- D-(+)-glucose monohydrate (Sigma-Aldrich, catalog number: 49159 )
- L-arginine-13C6,15N4 hydrochloride (Sigma-Aldrich, catalog number: 608033 )
- 1x Vogel’s salt (Vogel, 1956; Vogel, 1964)
- D-biotin (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: B1595 )
- Sucrose (Sigma-Aldrich, catalog number: S0389 )
- KCl (Thermo Fisher Scientific, AmbionTM, catalog number: AM9640G )
- NaCl (Thermo Fisher Scientific, InvitrogenTM, catalog number: 24740011 )
- Tris hydrochloride (Tris-HCl) (Sigma-Aldrich, catalog number: 10812846001 )
- MgCl2 (Thermo Fisher Scientific, AmbionTM, catalog number: AM9530G )
- Calcium chloride (CaCl2) (Sigma-Aldrich, catalog number: 793639-500G )
- NP-40 (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 85124 )
- Dithiothreitol (DTT) (Thermo Fisher Scientific, Thermo FisherTM, catalog number: 20290 )
- Spermidine (Sigma-Aldrich, catalog number: S0266 )
- HEPES (Sigma-Aldrich, catalog number: H4034 )
- EGTA (Sigma-Aldrich, catalog number: E3889 )
- Sodium dodecyl sulfate (SDS) (Sigma-Aldrich, catalog number: L3771 )
- EDTA (Thermo Fisher Scientific, Thermo FisherTM, catalog number: 17892 )
- Glycerol (Sigma-Aldrich, catalog number: G6279 )
- Glycogen, molecular biology grade (Thermo Fisher Scientific, Thermo FisherTM, catalog number: R0561 )
- Ethanol
- Paraformaldehyde (Sigma-Aldrich, catalog number: P6148 )
- Glycine (Thermo Fisher Scientific, InvitrogenTM, catalog number: 15527013 )
- Nuclease free H2O (Thermo Fisher Scientific, Thermo FisherTM, catalog number: R0581 )
- Liquid nitrogen
- Agarose (Thermo Fisher Scientific, Fisher ScientificTM, catalog number: BP1356100 )
- MNase (Sigma-Aldrich, catalog number: N3755-500 )
- RNase cocktail enzyme mix (Thermo Fisher Scientific, InvitrogenTM, catalog number: AM2286 )
- Proteinase K solution (Thermo Fisher Scientific, InvitrogenTM, catalog number: AM2548 )
- Wizard® SV gel and PCR clean-up system (Promega Corporation, catalog number: A9281 )
- EDTA-free protease inhibitor tablets (Roche Diagnostics, catalog number: 05892791001 )
- C6H8O7
- ZnSO4.7H2O
- Fe(NH4)2(SO4)2.6H2O
- CuSO4.5H2O
- MnSO4.1H2O
- H3BO3 (anhydrous)
- Na2MoO4.2H2O
- Chloroform
- MilliQ water
- Na3C6H5O7
- KH2PO4
- NH4NO3
- MgSO4.7H2O
- CaCl2.2H2O
- Liquid medium (see Recipes)
- Trace element solution (see Recipes)
- 50x Vogel’s salt (see Recipes)
- Lysis buffer (see Recipes)
- MNase resuspension buffer (see Recipes)
- Stop buffer (see Recipes)
- TE buffer (see Recipes)
Equipment
- Centrifuge
- Vortex
- Mortar and pestle
- Thermo-mixer (Eppendorf AG, model: Thermo-mixer R )
- Buhner funnel (Thermo Fisher Scientific, Fisher ScientificTM, catalog number: FB966B )
- Shaking incubator
- Agarose gel electrophoresis
Procedure
- Inoculate ~ 2 x 108 conidia of Neurospora crassa to 200 ml liquid medium in a 500 ml flask.
- Grow the culture with 100 rpm shaking under desired light and temperature conditions. Representative cultures were grown for three days at 24 °C and at 80 µE light intensity.
- In order to crosslink, add paraformaldehyde to the culture to a final concentration of 1% and incubate for 10 min shaking at 24 °C.
- Quench paraformaldehyde for 5 min with glycine at final concentration of 125 mM.
- Filter liquid medium through Buhner funnel and rinse mycelia with 500 ml of water (room temperature). Squeeze mycelia between tissue paper to get rid of all water and then place the mycelia in Eppendorf tube and immediately freeze in liquid nitrogen. At this step mycelia can be stored at -80 °C.
- Cool mortar and pestle by submerging in liquid nitrogen for 5 min. Take the mortar and pestle out and then place frozen mycelia immediately into mortar and grind mycelia with pestle until it is finely ground powder. At this step one should avoid warming mycelia/ground mycelia powder.
- Resuspend ~400 mg ground mycelia in 3.75 ml of lysis buffer with freshly added 0.5 mM DTT, 0.5 mM spermidine and EDTA-free protease inhibitor tablets.
- Mix the suspension well by vortexing and incubate on ice for 5 min.
- Distribute the suspension into five 1.5 ml Eppendorf tubes (750 μl per tube).
- Resuspend the MNase powder in 850 μl of MNase resuspension buffer (estimated concentration=0.58 U/μl). Aliquots can be stored at -20 °C. Avoid freeze-and-thaw cycles.
- Add respectively 0 [control], 0.75, 1.5, 3, 6 Units of MNase to each tubes. Incubate all samples at 25 °C for 1 h while shaking at 400 rpm in a Thermo-mixer.
- Add 85 μl of stop buffer to each sample to stop digestion and centrifuge at 20,000 x g for 20 min to pellet the cell debris. Transfer supernatants to new tubes
- Add 2 μl of RNase cocktail enzyme mix to each supernatant and incubate at 37 °C for 45 min shaking at 400 rpm in a Thermo-mixer.
- Add 15 μl of Proteinase K solution to each sample and incubate at 65 °C for 2 h shaking at 400 rpm in a Thermo-mixer.
- Add 40 μg glycogen and 1,250 μl of 100 % EtOH to each sample and incubate at -20 °C for 30 min.
- Perform centrifugation at max speed (~20,000 x g) for 30 min at 4 °C. Discard supernatant. Wash the DNA pellet with 700 μl 70 % ice-cold ethanol. Remove the ethanol slowly without disturbing the pellet. Drying the pellet is not required.
- Dissolve the washed DNA pellet in 100 μl DNAse/RNAse free H2O.
- Further purify DNA samples by using PCR-clean up kit.
- Load samples on 1.7% agarose gel (80 volts for 1 h) to analyze nucleosomal ladder (Figure1).
Representative data
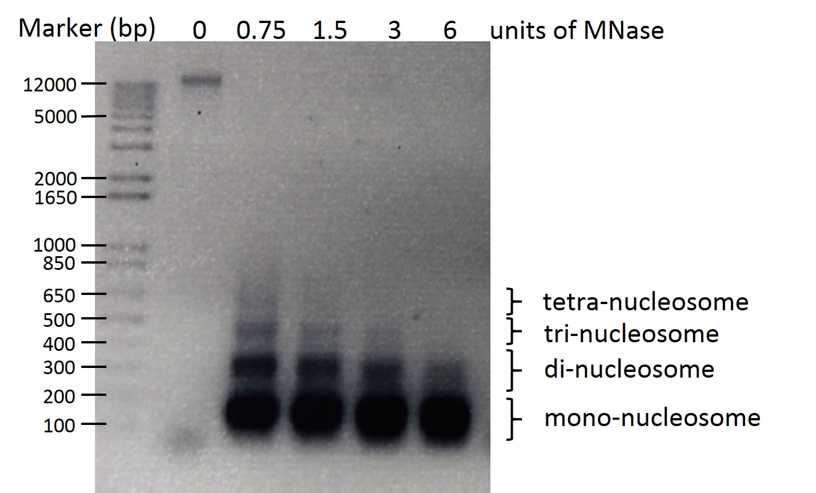
Figure 1. Representative image of MNase digested and purified Neurospora nucleosomal DNA loaded on 1.7% agarose gel
Recipes
- Liquid medium (prepare freshly)
2% (w/v) glucose monohydrate
0.5 % (w/v) L-arginine
1x Vogel’s salt (Vogel, 1956; Vogel, 1964)
10 ng/ml biotin - Trace element solution (stored at RT)
Dissolve indicated amount in 95 ml MilliQ water
5 g C6H8O7
5 g ZnSO4.7H2O
1 g Fe(NH4)2(SO4)2.6H2O
0.25 g CuSO4.5H2O
0.05 g MnSO4.1H2O
0.05 g H3BO3 (anhydrous)
0.05 g Na2MoO4.2H2O
Add 1 ml chloroform as preservative - 1 L 50x Vogel’s salt (stored at RT)
Dissolve indicated amount in 755 ml MilliQ water
125 g Na3C6H5O7
250 g KH2PO4
100 g NH4NO3
10 g MgSO4.7H2O
5 g CaCl2.2H2O
5 ml of trace element solution
250 µg biotin
No pH adjustment is necessary
Add 5 ml chloroform as preservative - Lysis buffer (stored at 4 °C)
250 mM sucrose
60 mM KCl
15 mM NaCl
15 mM Tris-HCl (pH 7.4)
3 mM MgCl2
1 mM CaCl2
0.2% NP-40 - MNase resuspension buffer (stored at 4 °C)
10 mM HEPES-KOH (pH 7.6)
50 mM KCl
1.5 mM MgCl2
0.5 mM EGTA
10% glycerol - Stop buffer (stored at RT)
2% SDS
100 mM EDTA (pH 8) - TE buffer (stored at 4 °C)
10 mM Tris-HCl (pH 8)
1 mM EDTA
Acknowledgments
This protocol was used for the work previously published in PLoS Genetics (Sancar et al., 2015). This work was supported by grants of the Deutsche Forschungsgemeinschaft: GRK1188 and SFB1036.
References
- Sancar, C., Ha, N., Yilmaz, R., Tesorero, R., Fisher, T., Brunner, M. and Sancar, G. (2015). Combinatorial control of light induced chromatin remodeling and gene activation in Neurospora. PLoS Genet 11(3): e1005105.
- Vogel, H. J. (1956). A convenient growth medium for Neurospora (Medium N). Microbiol Genet Bull 13:42-43.
- Vogel, H. J. (1964). Distribution of lysine pathways among fungi: Evolutionary implications. Am Nat 98: 435-446.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Sancar, C., Sancar, G. and Brunner, M. (2016). MNase Digestion for Nucleosome Mapping in Neurospora. Bio-protocol 6(11): e1825. DOI: 10.21769/BioProtoc.1825.
Category
Microbiology > Microbial genetics > DNA > Chromosomal
Microbiology > Microbial biochemistry > DNA
Molecular Biology > DNA > DNA-protein interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









