- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Assay of Arabinofuranosidase Activity in Maize Roots
Published: Vol 6, Iss 6, Mar 20, 2016 DOI: 10.21769/BioProtoc.1764 Views: 9727
Reviewed by: Zhaohui LiuDušan VeličkovićAgnieszka Zienkiewicz

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Enzymatic Starch Quantification in Developing Flower Primordia of Sweet Cherry
Nestor Santolaria [...] Afif Hedhly
Apr 5, 2025 1885 Views

New Approach to Detect and Isolate Rhamnogalacturonan-II in Arabidopsis thaliana Seed Mucilage
Dayan Sanhueza and Susana Saez-Aguayo
Sep 5, 2025 1240 Views

Detailed Method for the Purification of Rhamnogalacturonan-I (RG-I) in Arabidopsis thaliana
Liang Zhang [...] Breeanna R. Urbanowicz
Feb 5, 2026 99 Views
Abstract
Root is a perfect model for studying the mechanisms of plant cell growth. Along the root length, several zones where cells are at different stages of development can be visualized (Figure 1). The dissection of the root on these zones allows the investigation of biochemical and genetic aspects of different growth steps. Maize primary root is much more massive than the root of other Monocots and thus more convenient for such type of research. Plant cell wall, mainly consisting of polysaccharides, plays an important role in plant life. Therefore, measurement of plant carbohydrate content and glycoside-modifying enzyme activity in plant cells has become an important aspect in plant physiology. One of the well-documented changes of hemicelluloses molecules during elongation growth of monocots cells is the decrease of arabinose substitution of glucuronoarabinoxylans. This might be caused by changes in synthesis of this polysaccharide or by the action of arabinofuranosidases. Here, we describe the protocol of spectrophotometric measuring of arabinofuranosidase activity in maize root by the rate of hydrolysis of chromogenic substrate (4-nitrophenyl α-L-arabinofuranoside).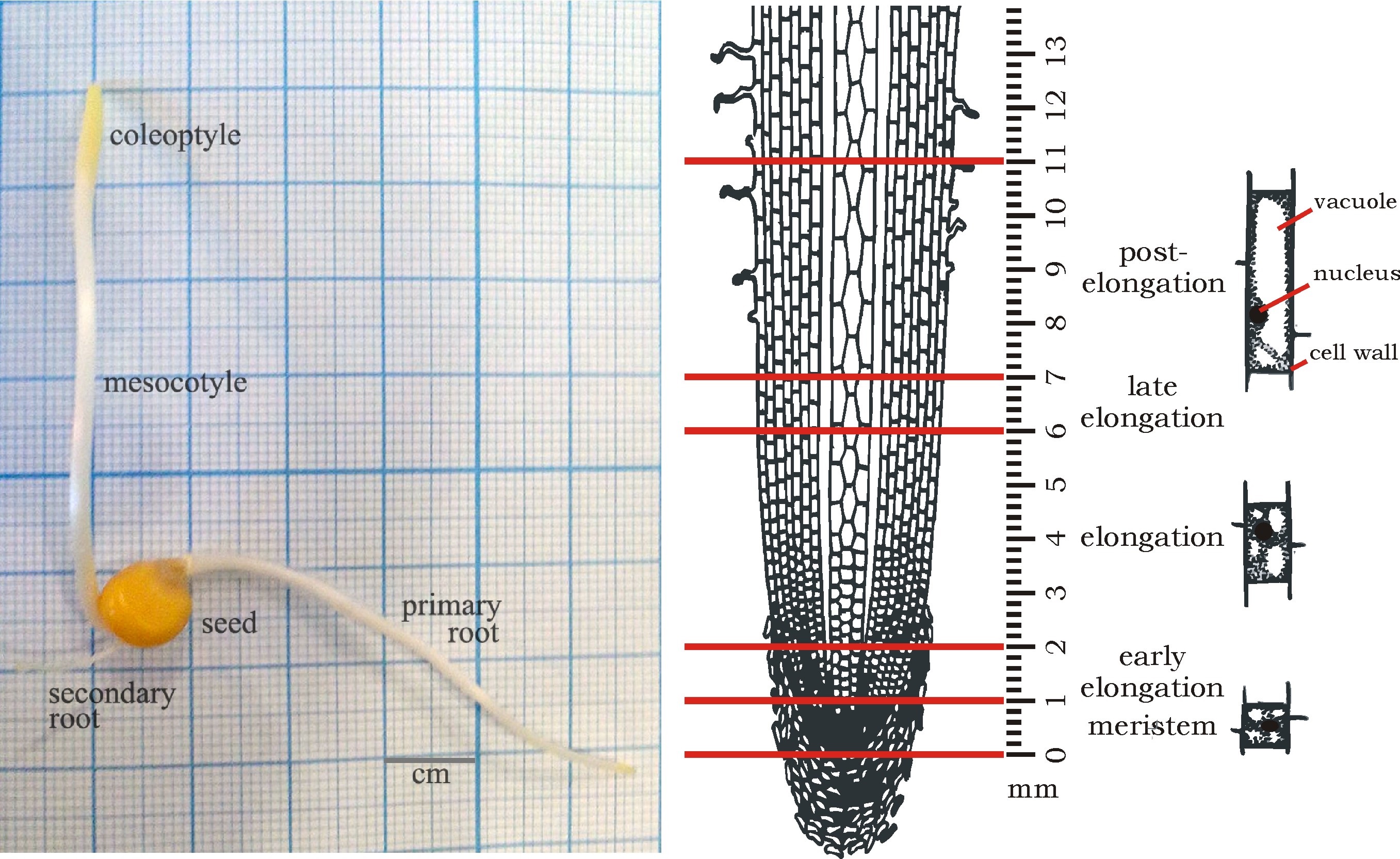
Figure 1. Scheme of plant material collection for further arabinofuranosidase assay. Four-day-old dark-grown maize seedling (left panel). Different zones of primary maize root and corresponding stages of cell development, according to Kozlova et al. (2012) (right panel).
Materials and Reagents
- Microcentrifuge tubes 1.5-2 ml
- CarboPac PA1 analytical column (4 x 250 mm) (Thermo Fisher Scientific, catalog number: 035391 )
- CarboPac PA1 guard column (4 x 50 mm) (Thermo Fisher Scientific, catalog number: 043096 )
- Maize seedlings
- 4-nitrophenyl α-L-arabinofuranoside (4NPA) (Sigma-Aldrich, catalog number: N3641 )
- 4-nitrophenol (NP) (spectrophotometric grade) (Sigma-Aldrich, catalog number: 1048 )
- DL-Dithiothreitol (Sigma-Aldrich, catalog number: D0632 )
- Quick StartTM Bradford 1x Dye (Bio-Rad Laboratories, AbD Serotec®, catalog number: 5000205 )
- Sodium Acetate Anhydrous (NaOAc) (Sigma-Aldrich, catalog number: W302406 )
- Sodium hydroxide (NaOH) (Sigma-Aldrich, catalog number: 306576 )
- L-(+)-ARABINOSE (Sigma-Aldrich, catalog number: W325501 )
- NaN3
- Na2CO3
- NaOCl
- Glacial acetic acid
- Ice
- MilliQ water (18 MΩ-cm)
- 5% (w/w) solution of NaOCl (see Recipes)
- 0.03% (w/w) NaN3 (see Recipes)
- 50 mM NaOAc buffer (pH 6.0) with 3 mM of dithiothreitol on 0.03% (w/w) NaN3 (see Recipes)
- 10 mM 4NPA (see Recipes)
- 1 M Na2CO3 (see Recipes)
- Calibration solutions for NP (see Recipes)
- 50% (w/w) NaOH solution (see Recipes)
- 15 mM NaOH (buffer A for HPAEC) (see Recipes)
- 1 M NaOAc in 0.1 M NaOH (buffer B for HPAEC) (see Recipes)
- Calibration solutions for arabinose (see Recipes)
Equipment
- Spectrophotometer (required wavelength is 405 nm)
- Thermostat (required temperature is 27 °C)
- Microcentrifuge (required spin rate is 10,000 x g)
- Thermoshaker (required temperature and spin rate are 37 °C and 600 rpm, correspondingly)
Note: Thermoshaker should be appropriate for incubation of microcentrifuge tubes. - Mini centrifuge/vortex
- Analytical balance with 0.1 mg readability
- Ion chromatography system DX-500 (Thermo Fisher Scientific, Dionex) equipped by electrochemical detector ED40, gradient pump GP40 and chromatography oven LC 30
- Refrigerator (required temperature is -20 °C)
- Automatic pipettes (100-1,000, 20-200 and 2-20 μl)
- Mortar and pestle
- Plastic grid and the tray of proper size
Note: The cell of grid should be approximately 1 cm2 or slightly bigger.
Software
- PeakNet software (Thermo Fisher Scientific, Dionex, model: version 5.21)
Procedure
Principal scheme of the experiment is shown in Figure 2.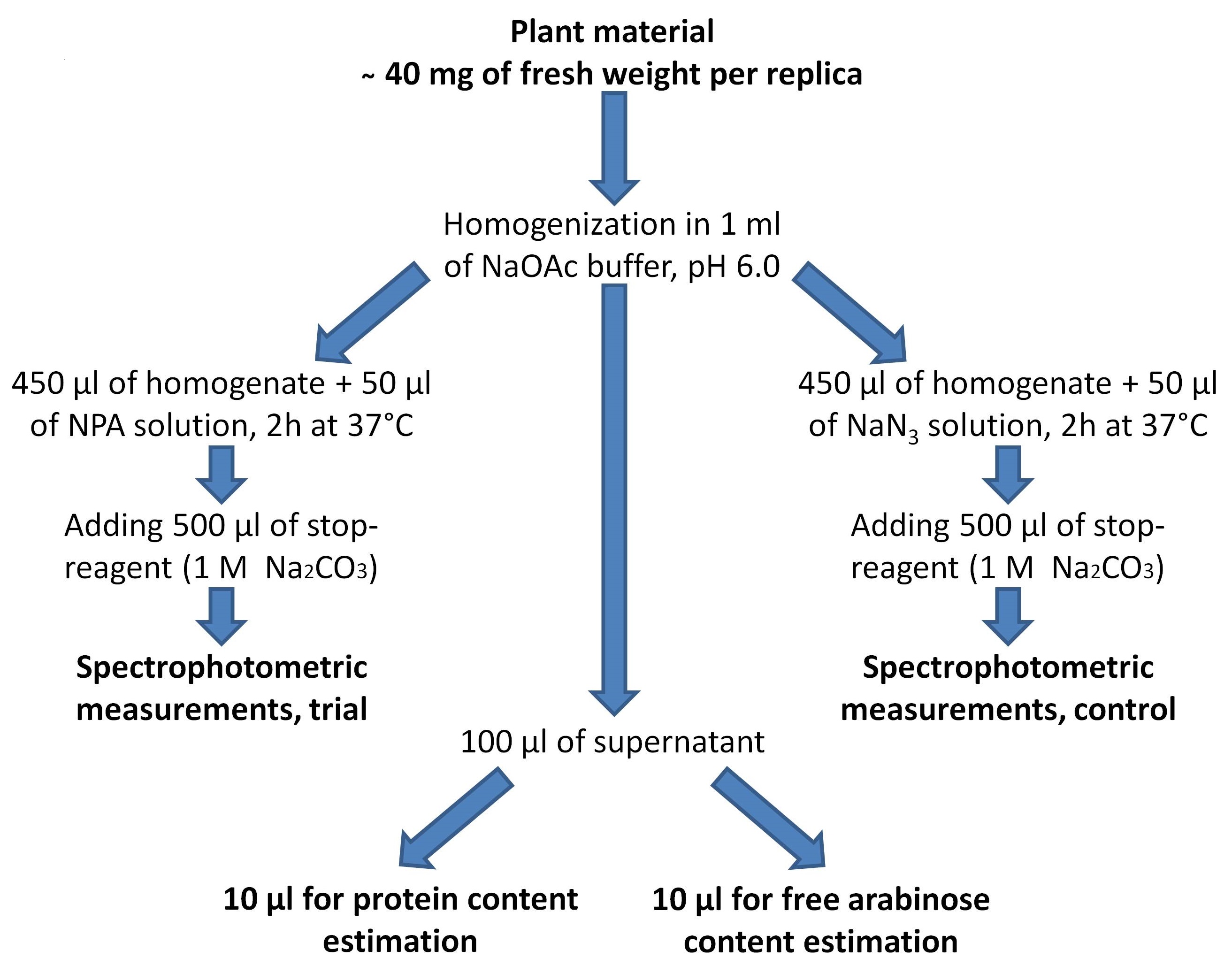
Figure 2. Scheme of the experiment
- Seeds of maize (Zea mays L.) were sterilized in 5% (w/w) solution of NaOCl for 10 min then were washed twice in water. Seeds were placed in the cells of plastic grid that was mounted in a tray with water. Plants were grown in the dark in a thermostat at 27 °C for 96 h. While any part of etiolated seedling can be used for the assay, we have used five zones (meristem, early elongation, elongation, late elongation, post-elongation) of primary roots (Figure 1). The recommended amount of plant material in each sample is 30-40 mg. One mm of maize root represents approximately one mg of fresh weight. Thus, root pieces of 10 mm length from three or four seedlings or pieces of 1-mm length from 30-40 seedlings are enough for one replica.
- 30-40 mg of fresh plant material was homogenized with mortar and pestle on ice in 1 ml of 50 mM NaOAc buffer (pH 6.0) with 3 mM of dithiothreitol to prevent the intramolecular and intermolecular disulfide bonds formation and with 0.03% (w/w) NaN3 to restrict bacterial contamination. Homogenate was transferred to microcentrifuge tube (1.5-2 ml). Homogenates that were already prepared could be stored in refrigerator (-20 °C) while other samples are being done.
- 1 ml of suspension was centrifuged for 10 min at 10,000 x g. Then 100 μl of supernatant were separated for further estimation of protein content according to Bradford (1976) (step 9) and free arabinose content estimation (step 10). Supernatants could be stored in refrigerator (-20 °C). Steps 9-10 could be carried out later. The arabinose and protein contents do not change if samples are frozen.
- Pellet was resuspended in the remaining 900 μl of buffer solution using Vortex. Plant material was not separated from supernatant because of the potential presence of wall-associated arabinofuranosidases that might be not extracted by the buffer solution. The suspension was divided into two equal parts (450 μl each) that were transferred to plastic tubes as control and trial variants of each sample.
- 50 μl of 10 mM 4NPA in 0.03% (w/w) NaN3 was added to the trial tubes, and 50 μl of 0.03% (w/w) NaN3 to the control tubes. Both of the variants were incubated in the thermoshaker at 600 rpm at 37 °C for 2 h. During that time cell wall arabinofuranosidases (either soluble or wall-associated) in trial tubes removed arabinose residues from 4NPA molecules producing yellow-colored NP. Enzymes in control tubes had no chromogenic substrate for reaction.
- The reaction was stopped by the addition of 500 μl of 1 M Na2CO3 both to the control and to the trial tubes. The coloration of the solution has become more pronounced at that moment due to increased pH (11.5).
- Samples were centrifuged for 10 min at 10,000 x g. Optical densities of both trial and control supernatants were measured on a spectrophotometer at wavelength 405 nm. The control mean was subtracted from the trial mean. The result was used for the determination of the amount of liberated NP according to calibration.
- Calibration measurements were done for NP solutions with concentrations in a range from 0.0025 mM to 0.25 mM (Figure 3). 500 μl of 1 M Na2CO3 were added to 500 μl of each NP solution, because it enhances the coloration.
- Determination of protein content.
- 490 μl of water was added to 10 μl aliquots of supernatant from step 3 of each sample. 500 μl of Bradford dye reagent was also added to each microcentrifuge tube and then mixed using vortex.
- After 5 min each sample was read on a spectrophotometer at 595 nm.
- Determination of free arabinose content.
- 90 μl of water waw added to 10 μl aliquots of supernatant from step 3 of each sample.
- 25 μl of obtained solution was directly injected to the column of HPAEC system without any pretreatment.
- 15 mM NaOH (buffer A) and 1 M NaOAc in 0.1 M NaOH (buffer B) were used as eluents. The column temperature was maintained at 30 °C, and the elution rate was 1 ml/min. The gradient elution was performed as follows: 0-20 min-100% A; 20-21 min-linear gradient until the A/B ratio = 90:10(%); 21-31 min-linear gradient until the A/B ratio = 70:30(%); 31-32 min-linear gradient until the A/B ratio 0:100(%); 32-42 min-100% B; 42-43 min-until the A/B ratio = 100:0(%). Column was equilibrated with eluent 100% A for 30 min.
- Arabinose content was determined by the area of corresponding peak using the PeakNet software and the calibration for arabinose standard.
- Calibration for free arabinose was done using solutions with concentrations in a range 2-10 μg/ml.
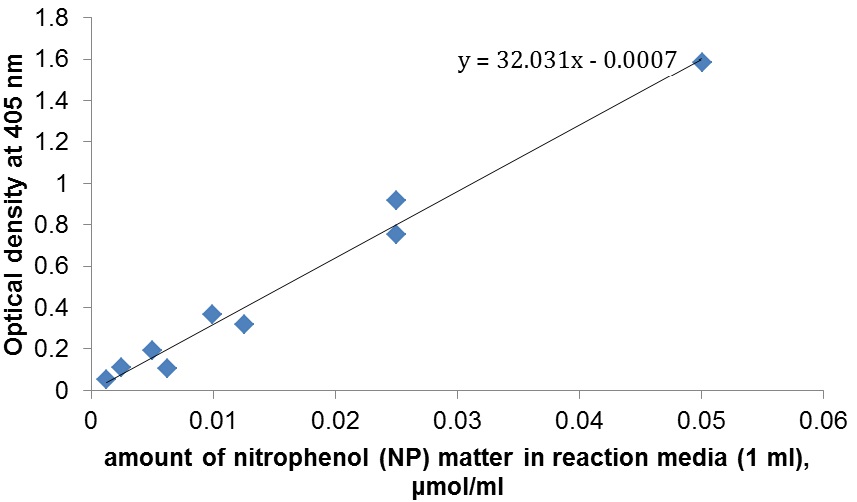
Figure 3. Calibration for different nitrophenol (NP) concentrations. Do not confuse the concentration and the amount of NP matter in reaction volume (see Table 1).
Notes
- Experiments should be repeated three-four times for each sample.
- One unit of arabinofuranosidase activity corresponds to the amount of enzyme that liberates one μmol of NP per minute. Differences of glycosyl-hydrolase activities between samples can be defined by using the units of enzymatic activity per mg of total proteins. Exact formula for arabinofuranosidase activity calculation is given in Figure 4.

Figure 4. Formula for calculation of arabinofuranosidases activity. The result will be obtained in milliUnits of activity per mg of protein content. - Described protocol with small modifications can be used for determination of any other glycosyl-hydrolase activity with synthetic aryl glycosides. Preliminary experiments may be useful for the optimization of incubation time and of plant material amount.
However, the enzyme activity that is measured only by the rate of chromogenic substrate hydrolysis can remain the potential one and can be not realized in plant tissues (Fry, 2004). The aryl-glycoside hydrolysis coupled with the demonstration of final product presence is much more convincing. Thus, steps 10 and 11 were included in protocol. If for any reason you cannot perform these stages, you may conclude only about potential activity of arabinofuranosidases.
Recipes
Note: The recipes that are given below let to produce the amount of solutions that is sufficient for calibration and four repeating of the experiments with five samples in each replica.
- 5% (w/w) solution of NaOCl
Dissolve 25 g of NaOCl in 475 ml of water - 0.03% (w/w) NaN3
Dissolve 0.03 g of NaN3 in 99.97 ml of water - 50 mM NaOAc buffer (pH 6.0) with 3 mM of dithiothreitol on 0.03% (w/w) NaN3
Dissolve 0.205 g of NaOAc and 0.0231 g of DL-dithiothreitol in 40 ml of 0.03% (w/w) NaN3
Adjust the pH to 6.0 by the adding of glacial acetic acid
Adjust the volume to 50 ml by the adding of 0.03% (w/w) NaN3 - 10 mM 4NPA
Dissolve 0.0027 g of 4NPA in 0.03% (w/w) NaN3
Adjust the volume of solution to 1 ml by additional 0.03% (w/w) NaN3 - 1 M Na2CO3
Dissolve 2.12 g of Na2CO3 in 0.03% (w/w) NaN3
Adjust the volume of solution to 20 ml by additional 0.03% (w/w) NaN3 - Calibration solutions for NP
Dissolve 0.0139 g of NP in 1 ml of 0.03% (w/w) NaN3 to produce 100 mM NP solution
Add 10 μl of 100 mM NP solution to 990 μl of 0.03% (w/w) NaN3 to produce 1 mM NP solution
Prepare 9 concentrations of NP according to the table below, place 500 μl of each calibration solution to individual tubes and add 500 μl of 1M Na2CO3 to all.
Table 1. NP solution preparation
NP concentration, mM Amount of NP matter in reaction volume (1 ml), μmol Volume of 1 mM NP solution, μl 0.03% (w/w) NaN3 solution volume, μl 0.25 0.125 250 750 0.1 0.05 100 900 0.05 0.025 50 950 0.025 0.0125 25 975 0.02 0.01 20 980 0.0125 0.00625 12.5 987.5 0.01 0.005 10 990 0.005 0.0025 5 995 0.0025 0.00125 2.5 997.5 - 50% (w/w) NaOH solution
HPAEC buffers should be prepared using template solution of NaOH
Add 5 ml of degasified MilliQ water (18 MΩ-cm) to 5 g of NaOH - 15 mM NaOH (buffer A for HPAEC)
Add 780 μl of 50% NaOH in 1,000 ml of degasified MilliQ water (18 MΩ-cm) - 1 M NaOAc in 0.1 M NaOH (buffer B for HPAEC)
Dissolve 82.05 g of NaOAc in degasified MilliQ water (18 MΩ-cm)
Adjust volume of solution to 1,000 ml by the adding of degasified MilliQ water (18 MΩ-cm)
Add 5.23 ml of 50% NaOHto NaOAc solution - Calibration solutions for arabinose
Dissolve 0.001 g of arabinose in 1 ml of MilliQ water to produce 10 mg/ml solution
Add 100 μl of 10 mg/ml arabinose solution to 900 μl of MilliQ water to obtain 1 mg/ml solution
Prepare 5 solutions of arabinose according to Table 2 below
Table 2. Arabinose solution preparationArabinose concentration, μg/ml Volume of 1 mg/ml arabinose solution, μl Volume of MilliQ, μl 2 2 998 3 3 997 5 5 995 8 8 992 10 10 990
Acknowledgments
This protocol was adapted from the previously published study (Kozlova et al., 2015). This work was partially supported by Russian Foundation for Basic Research (project ## 14-04-01002 and 15-04-02560).
References
- Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248-254.
- Fry, S. C. (2004). Primary cell wall metabolism: tracking the careers of wall polymers in living plant cells. New Phytol 161:641-675.
- Kozlova, L. V., Gorshkov, O. V., Mokshina, N. E. and Gorshkova, T. A. (2015). Differential expression of alpha-L-arabinofuranosidases during maize (Zea mays L.) root elongation. Planta 241(5): 1159-1172.
- Kozlova, L. V., Snegireva A. V. and Gorshkova T. A. (2012). Distribution and structure of mixed linkage glucan at different stages of elongation of maize root сells. Russ J Plant Phys 59: 339-347.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Kozlova, L. V., Mikshina, P. V. and Gorshkova, T. A. (2016). Assay of Arabinofuranosidase Activity in Maize Roots. Bio-protocol 6(6): e1764. DOI: 10.21769/BioProtoc.1764.
Category
Plant Science > Plant biochemistry > Carbohydrate
Biochemistry > Carbohydrate > Polysaccharide
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










