- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Laser Microirradiation and Temporal Analysis of XRCC1 Recruitment to Single-strand DNA Breaks
Published: Vol 6, Iss 5, Mar 5, 2016 DOI: 10.21769/BioProtoc.1746 Views: 9210
Reviewed by: Nicoletta CordaniAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Targeted Nucleotide Substitution in Mammalian Cell by Target-AID
Takayuki Arazoe [...] Akihiko Kondo
Jun 5, 2017 19754 Views

Detecting the Activation of Endogenous Small GTPases via Fluorescent Signals Utilizing a Split mNeonGreen: Small GTPase ActIvitY ANalyzing (SAIYAN) System
Miharu Maeda and Kota Saito
Jan 5, 2026 483 Views

A Quantitative DNA Fiber Assay to Monitor Replication Fork Progression, Protection, and Restart
Debanjali Bhattacharya and Ganesh Nagaraju
Feb 5, 2026 146 Views
Abstract
The DNA molecule is exposed to a multitude of damaging agents that can compromise its integrity: single (SSB) and double strand breaks (DSB), intra- or inter-strand crosslinks, base loss or modification, etc. Many different DNA repair pathways coexist in the cell to ensure the stability of the DNA molecule. The nature of the DNA lesion will determine which set of proteins are needed to reconstitute the intact double stranded DNA molecule. Multiple and sequential enzymatic activities are required and the proteins responsible for those activities not only need to find the lesion to be repaired among the millions and millions of intact base pairs that form the genomic DNA but their activities have to be orchestrated to avoid the accumulation of toxic repair intermediates. For example, in the repair of Single Strand Breaks (SSB) the proteins PARP1, XRCC1, Polymerase Beta and Ligase III will be required and their activities coordinated to ensure the correct repair of the damage.
Furthermore, the DNA is not free in the nucleus but organized in the chromatin with different compaction levels. DNA repair proteins have therefore to deal with this nuclear organization to ensure an efficient DNA repair. A way to study the distribution of DNA repair proteins in the nucleus after damage induction is the use of the laser microirradiation with which a particular type of DNA damage can be induced in a localized region of the cell nucleus. The wavelength and the intensity of the laser used will determine the predominant type of damage that is induced. It is important to note that other lesions can also be generated at the microirradiated site.
Living cells transfected with the fluorescent protein XRCC1-GFP are micro-irradiated under a confocal microscope and the kinetics of recruitment of the fluorescent protein is followed during 1 min. In our protocol the 405 nm laser is used to induce SSB.
Materials and Reagents
- Glass bottom 35 mm Petri dishes (MatTek, catalog number: P35G-1.5-20-C )
- HeLa adherent cells in culture
- Plasmids coding for the fluorescent protein XRCC1-GFP
Note: The XRCC1 sequence (NCBI P18887) was cloned in the Clontech pEGFP-N1 Vector at the restriction sites EcoRI/ApaI. - Transfection agents (LipoFectamine 2000) (Life Technologies, catalog number: 11668027 )
Note: Currently, it is “Thermo Fisher Scientific, Invitrogen™, catalog number: 11668027”. - DMEM [DMEM(1x) + GlutaMAX™ (Thermo Fisher Scientific, Gibco™, catalog number: 31966-021 )] containing 10% of fetal bovine serum (Thermo Fisher Scientific, Gibco™, catalog number: 16000-044 )
Equipment
- Cell incubator (Thermo Fisher Scientific, model: NAPCO 5400 )
Note: Currently, it is “BioSurplus, catalog number: NAPCO 5400 ”. - Nikon A1 inverted confocal microscope equipped with an environmental chamber allowing the control of temperature, humidity and gas mixture
Note: The lasers used are the following: 405 nm laser for the micro-irradiation, and a 488 nm Argon-laser to visualize the GFP fluorescence. The objective 63x was used.
Software
- The NIS Software
Note: It was used for quantification of the fluorescence intensity at the microirradiation region during time, using the Time Measurement function.
Procedure
- HeLa cells were seeded in 35 mm glass bottom Petri dishes (250,000 cells per dish) and transfected 24 h later with the XRCC1-GFP plasmid using Lipofectamine 2000 according to manufacturer recommendations.
- 24 h after transfection, microirradiation was carried out with a 405 nm diode laser set to 5% power. The laser power at the exit of the fiber was of 4 mW, and around 1 mW watts at the exit of the 10x Obj.
- Measurements were performed by immobilizing the laser at 100% in a point bleach with the Digital Handheld Optical Power PM100D from THORLABS.
- Stimulation and acquisition were performed with the 60x objective at a zoom of 4 using an image size of 512 x 512.
- A stimulation line of 5 µm was defined and microirradiation performed for 6 sec. Six images were taken before micro-irradiation, to calculate the basal level of fluorescence. After micro- irradiation, an image was taken every 2 sec during 1 min.
- Between 10 and 20 cells are micro-irradiated in each experiment.
Analysis and quantification
- The fluorescence intensity at the micro-irradiated region is measured for each time point. Six images are taken before the micro-irradiation in order to quantify a mean of fluorescence that will be considered as the basal level of the protein in the region and used to normalize the measurements (this value is set to 1).
- In order to quantify the enrichment factor of XRCC1 in the micro-irradiation region the intensity observed for each time point is divided by the mean intensity measured before the micro-irradiation.
- The mean of at least 10 cells is displayed in the graph. Error bars represent the SEM.
- It is important to check if there is bleaching during the acquisition process and correct for that if necessary. In order to do that we define a Standard Region of Interest (ROI-2) in another cell expressing similar amounts of XRCC1-GFP that is in the same scan area. Fluorescence intensity is monitored both in the micro-irradiated cell and in the non micro-irradiated one during both the stimulation and the acquisition period. If there is bleaching, we will observe a decrease in the fluorescence in the non micro-irradiated cell. It was not the case in our system, so no further corrections were required. If bleaching is observed, it is recommended to adjust the acquisition conditions in order to reduce that problem.
Representative data
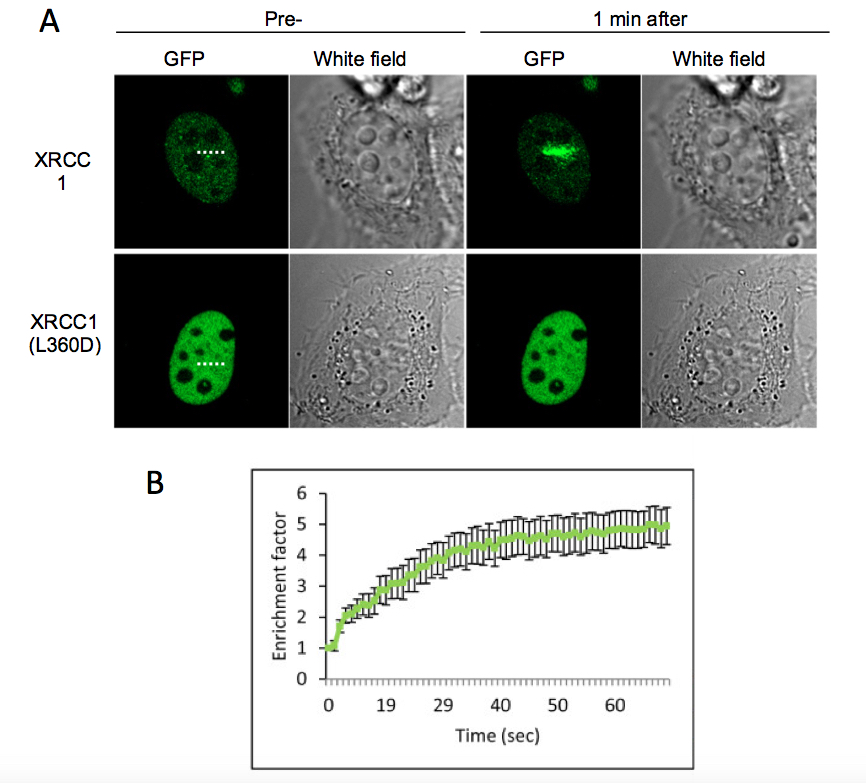
Figure 1. HeLa cell expressing the XRCC1-GFP or the mutant protein XRCC1(L360D) were microirradiated with a 405 nm laser. A) Presented images correspond to the cell before microirradiation and 1 min after microirradiation. The microirradiation region is indicated by a dashed line. The mutant protein XRCC1(L360D), that has not any more the ability to be recruited to SSB was used as a control. B) Fluorescence intensity corresponding to XRCC1-GFP at the microirradiated region was quantified and displayed in the graph. Error bars represent the SEM of 10 independent cells.
Acknowledgments
We thank the IRCM microscopy facility. This work was funded by INSERM and grants from the Association pour la Recherche sur le Cancer (PJA 20131200165) and the CEA Radiobiology program.
References
- Campalans, A., Kortulewski, T., Amouroux, R., Menoni, H., Vermeulen, W. and Radicella, J. P. (2013). Distinct spatiotemporal patterns and PARP dependence of XRCC1 recruitment to single-strand break and base excision repair. Nucleic Acids Res 41(5): 3115-3129.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Campalans, A. and Radicella, J. P. (2016). Laser Microirradiation and Temporal Analysis of XRCC1 Recruitment to Single-strand DNA Breaks. Bio-protocol 6(5): e1746. DOI: 10.21769/BioProtoc.1746.
Category
Cancer Biology > General technique > Genetics > DNA damage
Cell Biology > Cell imaging > Confocal microscopy
Molecular Biology > DNA > DNA damage and repair
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











