- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Stable Isotope Resolved Metabolomics Studies in ex vivo TIssue Slices
Published: Vol 6, Iss 3, Feb 5, 2016 DOI: 10.21769/BioProtoc.1730 Views: 11721
Reviewed by: Masahiro MoritaJustine MarsolierAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Hypoxia Studies with Pimonidazole in vivo
Kristina Y. Aguilera and Rolf A. Brekken
Oct 5, 2014 28611 Views

13C Tracer Studies of Metabolism in Mouse Tumor Xenografts
Andrew N. Lane [...] Teresa W-M. Fan
Nov 20, 2015 12868 Views

Analysis of Leukemia Cell Metabolism through Stable Isotope Tracing in Mice
Nick van Gastel [...] David T. Scadden
Oct 5, 2021 5228 Views
Abstract
An important component of this methodology is to assess the role of the tumor microenvironment on tumor growth and survival. To tackle this problem, we have adapted the original approach of Warburg (Warburg, 1923), by combining thin tissue slices with Stable Isotope Resolved Metabolomics (SIRM) to determine detailed metabolic activity of human tissues. SIRM enables the tracing of metabolic transformations of source molecules such as glucose or glutamine over defined time periods, and is a requirement for detailed pathway tracing and flux analysis. In our approach, we maintain freshly resected tissue slices (both cancerous and non- cancerous from the same organ of the same subject) in cell culture media, and treat with appropriate stable isotope-enriched nutrients, e.g., 13C6-glucose or 13C5, 15N2-glutamine. These slices are viable for at least 24 h, and make it possible to eliminate systemic influence on the target tissue metabolism while maintaining the original 3D cellular architecture. It is therefore an excellent pre-clinical platform for assessing the effect of therapeutic agents on target tissue metabolism and their therapeutic efficacy on individual patients (Xie et al., 2014; Sellers et al., 2015).
Keywords: Tissue slicesMaterials and Reagents
- 25 ml T Flasks NC vent cap (SARSTEDT AG & Co, catalog number: 83.1810.002 )
- Portable container for liquid nitrogen (Nalgene plastic dewar) (Thermo Fisher Scientific, catalog number: S34074C )
- Sterile syringes and needles (Thermo Fisher Scientific, catalog number: 10142534 )
- Disposable transfer pipets (Samco fine tip, 1 ml) (VWR International, catalog number: 16001192 )
- Aerosol barrier tips for 1 ml and 1-200 μl (Thermo Fisher Scientific, catalog number: 02-707-42 )
- Screw cap plastic vials (2 ml) color coded caps (yellow, blue, green and red) (USA Scientific, catalog number: 1420-8706 , 1420-8701 , 1420-8702 and 1420-9704 )
- Snap top plastic vials (1.5 ml) (USA Scientific, catalog number: 1615-5510 )
- 15 ml Falcon tubes (SARSTEDT AG & Co, catalog number: 62.554.205 )
- Dialyzed, sterile filtered fetal bovine serum (FBS) (free of serum metabolites),10-12 kDa (Atlanta Biochemical, catalog number: S12650 )
- Tracer examples: 13C6-glucose, 13C2-1, 2-glucose, 13C5,15N2-glutamine
- Sources: 13C6-glucose/D-glucose ([U-13C], 99%) (Cambridge Isotope Laboratories, catalog number: CLM-1396-CTM), 13C2-1, 2 glucose/D-glucose (1, 2-13C2, 99%) (Cambridge Isotope Laboratories, catalog number: CLM-504), 13C5, 15N2-glutamine/ L-glutamine (13C5, 99%; 15N2, 99%) (Cambridge Isotope Laboratories, catalog number: CNLM-1275 ) or
Isotec: D-13C6-glucose (Sigma-Aldrich, catalog number: 660663 ), 13C2-1, 2 glucose (Sigma-Aldrich, catalog number: 661422 ), L-Glutamine-13C5, 15N2 (Sigma-Aldrich, catalog number: 607983 ) - Penicillin + Streptomycin: GE Healthcare PEN/STREP/FUNGIZONE 100 ml (Thermo Fisher Scientific, catalog number: SV3007901 )
- ProtocolTM 10% Neutral buffered formalin (Thermo Fisher Scientific, catalog number: 032-059 )
- 25% (w/v) sterile filtered 13C glucose (0.2 μm) in PBS (Stock solution can be frozen, aliquoted, and stored at 4 °C)
- Liquid nitrogen
- 70% ethanol (v/v)
- 60% acetonitrile in water (v/v) (Sigma-Aldrich, catalog number: L010400 )
- Sodium chloride (NaCl) (Thermo Fisher Scientific, catalog number: S271-1 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9541 )
- Sodium phosphate dibasic (Na2HPO4) (Sigma-Aldrich, catalog number: S0876 )
- Potassium phosphate monobasic (KH2PO4) (Sigma-Aldrich, catalog number: P9791 )
- Amino acids: Glycine, L-Arginine, L-Asparagine, L-Aspartic acid, L-Cystine 2HCl, L-Glutamic Acid, L-Glutamine, L-Histidine, L-Hydroxyproline, L-Isoleucine, L-Leucine, L-Lysine hydrochloride, L-Methionine, L-Phenylalanine, L-Proline, L-Serine, L-Threonine, L-Tryptophan, L-Tyrosine disodium salt dehydrate, L-Valine
- Vitamins: Biotin, Choline chloride, D-Calcium pantothenate, Folic Acid, i-Inositol, Niacinamide, Para-Aminobenzoic Acid, Pyridoxine hydrochloride, Riboflavin, Thiamine hydrochloride, Vitamin B12
- Calcium nitrate [Ca(NO3)2.4H2O]
- Magnesium sulfate (MgSO4)
- Glutathione (reduced)
- Phenol Red
- Relevant medium (e.g. DMEM, RPMI, other defined medium) which lacks the tracer of interest:
a.Dulbecco′s Modified Eagle′s Medium (DMEM) is a powder formula, free of glucose, glutamine, pyruvate bicarbonate, and phenol red, giving considerable flexibility in formulation for SIRM studies (Sigma-Aldrich, catalog number: D5030 ) (see Recipes)
b.RPMI 1640 is a liquid medium free of glucose and glutamine, but contains bicarbonate and phenol red (MP Biomedicals, catalog number: 091646854 ) (see Recipes) - 0.2 µm sterile filtered Phosphate Buffered Saline (PBS) (see Recipes)
- Medium composition for 0.2% 13C6-glucose, 2 mM 12C-Gln (100 ml) (see Recipes)
- Medium composition for 0.2% 12C glucose, 2 mM 13C5, 15N2-Gln (100 ml) (see Recipes)
Equipment
- Class II Biosafety Hood
- Trigas incubator with oxygen sensor and CO2 sensor (Thermo Fisher Scientific, model: Hera cell 150i )
- Sterilized rocker (Rotoshake Genie) (Scientific Industries, model: SI-1100 )
- Liquid nitrogen freezer for storage
- K2-EDTA vacutainers (“purple top”) (BD, catalog number: 366643 )
- Refrigerated centrifuge with swing out rotor that can accept vacutainers [e.g. Sorvall Legend X1R (Thermo Fisher Scientific, catalog number: 75-004-261 ) with a rotor (Thermo Fisher Scientific, catalog number: 75003181 )]
- Pipettors (variable size ranges) (USA Scientific ErgoOne)
- Weck Knife/Dermatome (George Tiemann & Co., catalog number: 222-5-523 )
- Weigh boats (Thermo Fisher Scientific, catalog number: 08732113 and 08732115 )
- 4-place balance (Thermo Fisher Scientific, Mettler-Toledo, catalog number: 0133525 )
- Ice bucket (Thermo Fisher Scientific, catalog number: 02-591-44 )
- Sharp dissecting scissors (Thermo Fisher Scientific, catalog number: 08940 )
- Excelta™ Plastic Tweezers (Thermo Fisher Scientific, catalog number: 17-456-066 )
- Digital camera
Procedure
- Tissue procurement
All tissue must be procured under an IRB approved protocol. As live human tissue is handled, all personnel must undergo and maintain biosafety, HIPAA and CITI certifications.
An overview of the whole process is given in Scheme 1.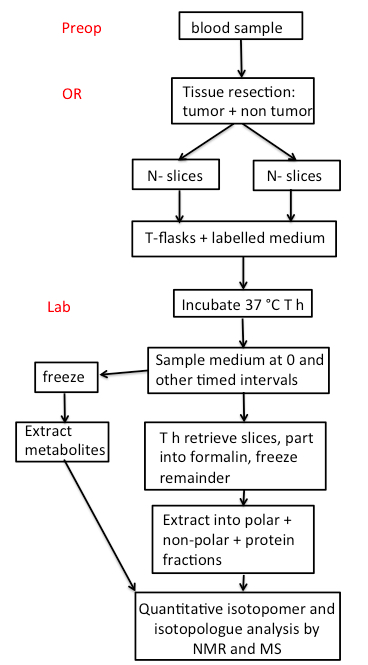
Scheme 1. FlowChart. Preop: Preoperative room; OR: Operating Room. The number of thin slices to be taken depends on the size of the tumor. A piece of tissue is also flash frozen in the OR, and additional tissue is placed in formalin for pathological analysis.- Blood samples provide overall information about the metabolic status of the individual subjects, and the buffy coat can be used for extracting DNA or RNA for sequence analysis.
- A 10 ml sample of blood should be drawn preoperatively into a purple top vacutainer (K2-EDTA) preoperatively. Other anticoagulants such as citrate or heparin should not be used as they interfere with metabolic assays. A blood sample should also be drawn perioperatively after resection. The blood is inverted twice to ensure dissolution of the EDTA, and kept on ice immediately after blood draw. The blood should be separated into packed red cells, buffy coat and plasma within 30 min by centrifuging at 3,500 x g for 15 min at 4 °C in a swing out rotor.
Subsequent operations should be carried out in a BSL2+ biosafety cabinet.
Note: We use the following color codes for storage: Red = whole blood, yellow = plasma, green = buffy coat, blue = urine. - Plasma is aspirated into prechilled sterile 2 ml screw cap vials at 1 ml aliquots and flash frozen in liquid N2. Buffy coat is aspirated using a wide mouth plastic pipette into a 2 ml screwcap vial and flash frozen in liquid N2.
- These experiments have been carried out on fresh slices of paired cancerous (CA) and non-cancerous (NC) lung tissues resected from non-small cell lung cancer (Xie et al., 2014) and pancreatic cancer patients. Upon resection, thin slices (0.5-1 mm thick) of tissue are excised from the surface of visually non-necrotic or fibrotic tumor regions using a Weck microtome in the Operating Room (OR), within approximately 5-10 min of resection. Roughly 1 cm2 tissue is targeted (see Figure 1 A). Control non-cancerous tissue from a distant (>10 cm) region is obtained similarly. A pathologist on-site inspects the CA and NC tissue specimens. Highly necrotic tissue is discarded.
- At the same time, a small piece of bulk CA tissue should be placed in DMEM or other appropriate medium kept room temperature for implantation into a recipient NSG mouse as patient-derived xenograft or PDX. A small piece each of CA and NC tissues is soaked in formalin for pathological examination or flash frozen in liquid N2 for image-based metabolic analysis.
- Where tissue acquisition in the OR is impractical (such as colorectal or breast cancer resections), the slices can be prepared in the pathology laboratory located close to the OR. For comparison with freshly resected tissue, speed is essential as metabolism is rapidly changing. Whenever feasible, tissue freezing should be performed in the OR.
- The slices are placed into a drop of sterile PBS on two sterilized weigh boats to prevent sticking and for spreading slices evenly. Each weight boat is pre-numbered for CA or NC slices. The tissues on weight boats are then photographed. Each slice is rinsed briefly with sterile PBS, blotted (twice) and then carefully placed into pre-numbered (using ethanol-resistant marker pen) pre-tared (tare weight recorded) T25-flasks containing 10 ml DMEM (or other relevant medium) with the appropriate tracer (e.g. 10 mM 13C6-glucose or 2 mM 13C5, 15N2-glutamine), 10% dialyzed FBS (as needed), and 1x penicillin + streptomycin. The flasks with slices are brought to the culture room as soon as possible and sprayed with 70% ethanol, and wiped dry before placing them in the Biosafety hood.
- Pipet 200 μl culture media from each flask into 1.5 ml snap-cap tubes (t0-time zero media samples). Centrifuge for 10 min at 10,000 x g at 4 °C to remove tissue debris. Transfer 100 μl to tared 1.5 μl snap-cap tube for metabolite extraction and weigh the media transferred. Transfer the remaining media to a 1.5 ml screw-cap tube for long-term storage at -80 °C.
- Weigh flasks in a 2-place balance inside the Biosafety hood and record weight on the flask.
- Transfer flasks to a CO2 incubator containing a rocker set to low amplitude rocking (sufficient to ensure that the liquid moves over the slices without non-laminar flow) Figure 1B) set to 37 °C and 5% CO2, with oxygen set to the desired level (e.g. 20%, 1%).
- The flasks are continuously and gently rocked for 24 to 48 h for gas exchange and to maintain constant nutrient supplies at the tissue surface, while avoiding local buildup of waste products such as acids.
- As needed, the medium can be refreshed every 12 h, and sampled at 0, 6, 12, 24…. h for analysis of nutrient uptake and waste production. The flasks are weighed before and after each medium sampling point and flask weights are recorded on the flasks.
- Blood samples provide overall information about the metabolic status of the individual subjects, and the buffy coat can be used for extracting DNA or RNA for sequence analysis.
- Tissue Harvesting
- After 24 h incubation, weigh flasks.
- Place flasks on ice immediately after removing from the incubator to minimize further metabolism. Up to 6 flasks can be harvested at a time. Keep tissue slices on ice as much as possible during harvest.
- Using a transfer pipet, aspirate and transfer the conditioned media into 15-ml conical centrifuge tubes.
- Centrifuge media for 15 min at 3,500 to 4,690 x g, 4 °C to remove any particulates and debris.
- Pipet 100 μl T24 media supernatant into 1.5-ml snap-cap tubes for metabolite extraction.
- Pipet 1 ml media aliquot into 2-ml screw-cap tubes for long-term storage at -80 °C.
- The remaining medium is stored separately in a 7 ml vial at -80 °C for purposes such as exosome isolation.
- Invert and tap the flask to move the tissue slices into the cap or neck region of the flask for retrieval. Keep flask inverted on ice.
- Wash tissue slices 3x consecutively in ice-cold 10 ml cold PBS each in a 50 ml beaker.
- Blot dry the tissue slices on Kimwipe and photograph the flattened slice on a small weigh boat.
- Weigh the whole tissue slice on small weigh boats and record the weight.
- Split a very small piece for preservation in 1 ml buffered formalin in a 1.5 ml snap-cap tube for histology. The remaining tissue slice is split evenly and each piece should weigh no more than 20-30 mg by wet weight to facilitate tissue homogenization and extraction efficiency. Immediately after weighing, each piece is flash-frozen in liq. N2 and placed in a pre-liq. N2 chilled 1.5-ml snap-cap tube for long-term storage at -80 °C.
- After 6-8 h in formalin, replace the formalin with 70% ethanol for the tissue pieces prepared for histology.
- Homogenize tissues in cold 60% acetonitrile (v/v) and extract tissue homogenates for metabolite analyses according to standardized protocols (Fan, 2012; Fan, 2010) before analyses using stable isotope-resolving analytical techniques (e.g. NMR and MS) (Fan, 2012) (Figure 1C) (Lane et al., 2008).
- This Protocol describes the procedure for stable isotope labeling of thin tissue slices. SIRM analysis involves the quantification of isotopomers (by NMR) and isotopologues (MS) that result from metabolic transformations of source molecules (e.g. 13C glucose or 13C, 15N Glutamine) in cells or tissue (Figure 1C). The techniques of SIRM analysis by NMR and mass spectrometry are described in detail in Sellers et al. (2015); Lane et al. (2008) and Fan et al. (2012).
- After 24 h incubation, weigh flasks.
Representative data
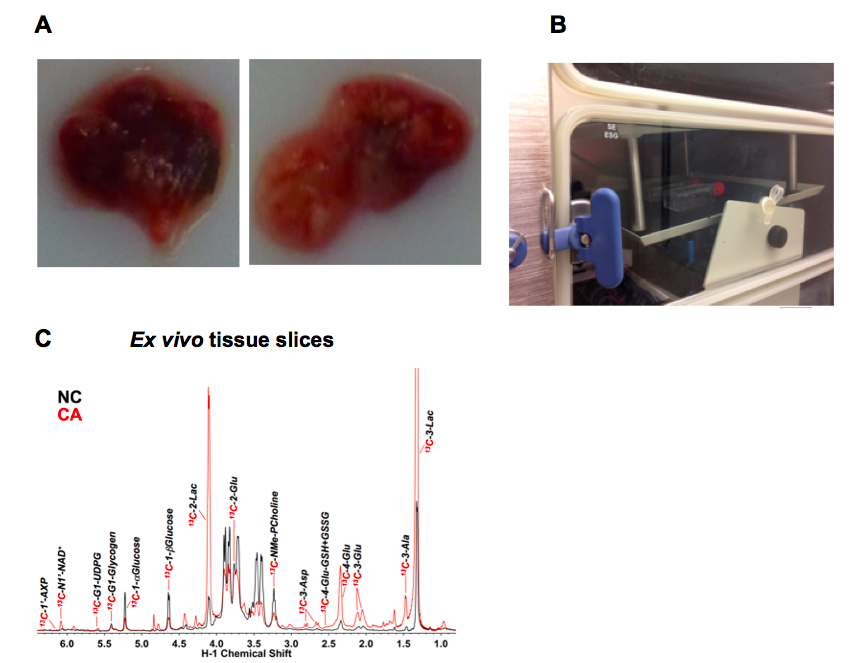
Figure 1 Example ex vivo tissue slice experiment. A. Example thin slices of non-cancerous lung tissue (NC, left) adjacent to a lung adenocarcinoma (CA, right); B. T25-flasks on a rocker inside a CO2 incubator; C. Representative 1D 1H{13C} HSQC NMR spectra (recorded at 14.1 T, 15 °C) of extracts of CA versus NC lung slices from an non small cell lung cancer (NSCLC) patient incubated for 24 h in the presence of 10 mM 13C6-glucose. The tissue slices were pulverized and extracted as described (Sellers et al., 2015; Fan, 2012) which produces three phases- an upper aqueous phase containing polar metabolites, a lower organic phase containing non-polar metabolites (mainly lipids) and an interfacial phase that contains protein. Here the upper phase was lyophilized and redissolved in a phosphate buffer containing 50% D2O and 25 nmol DSS-d6 that serves both as a chemical shift reference and a concentration standard (Fan and Lane, 2013). The HSQC spectrum detects protons attached directly to 13C, and thus gives a readout of the metabolites that have incorporated 13C from the source molecule (glucose in this instance). The spectra of cancer and non-cancerous tissues are recorded under identical conditions, and the absolute intensities are normalized to the tissue protein weight. Peak areas were determined using peak fitting functions in MNOVA (Mestrelab Research, Santiago de Compostela, Spain)
Enhanced production of various 13C labeled metabolites in the CA tissue slice is evident, including 13C-lactate (Lac), which is consistent with the Warburg effect or accelerated glycolysis in tumor tissues (Warburg, 1956).
Notes
- These procedures have been tested on freshly resected NSCLC and pancreatic cancer, as well as in patient derived mouse xenografts. Other tissues may need experimentation with the composition of the medium and length of incubation period for metabolic viability.
- Larger or inflammatory tumors may have substantial areas of necrosis or fibrosis that need to be avoided.
- A Weck microtome is hand held, but with practice the surgeons can reproducibly produce slices < 1 mm thick. Slice thickness is easily estimated by measuring the area of the slice from a photograph and from the wet weigh as the average thickness h = weight/area.
- Some tumors are highly mucilaginous and are more difficult to slice reproducibly.
- An alternative to a Weck microtome is a vibrating microtome, which can reproducibly generate thinner slices from firm but not soft tissues (cf10), but is much slower. Very thin slices (100-200 μm) may show a proportionately larger wounding response and are more fragile.
- As many tumors are heterogeneous not only in the cancer/stromal content in different regions of the tumor but also in terms of genetics, it is advisable to obtain multiple slices from the tumor to cover this heterogeneity. This also makes histopathological examination of each slice critically important.
- The margins of some tumors are not obvious without pathological examination. Tissue proximal to the tumor as well as distal from the tumor should be sampled for comparison.
Recipes
- DMEM and RPMI Medium 1640
For SIRM studies, the glutamine or glucose free version of the medium should be used, with supplementation of the appropriate concentration of 13C-enriched precursors in the base medium.COMPONENTS Molecular Weight Concentration (mg/L) Molarity (mM) RPMI Molarity (mM) DMEM Amino Acids Glycine 75 10 0.133 0.40 L-Arginine 174 200 1.15 0.483 L-Asparagine 132 50 0.379 - L-Aspartic acid 133 20 0.150 - L-Cystine 2HCl 313 65 0.208 0/0.2 L-Glutamic Acid 147 20 0.136 - L-Glutamine 146 300 2.05 2 L-Histidine 155 15 0.0968 0.27 L-Hydroxyproline 131 20 0.153 - L-Isoleucine 131 50 0.382 0.8 L-Leucine 131 50 0.382 0.8 L-Lysine hydrochloride 146 40 0.274 1.0 L-Methionine 149 15 0.101 0.2 L-Phenylalanine 165 15 0.0909 0.4 L-Proline 115 20 0.174 - L-Serine 105 30 0.286 0.4 L-Threonine 109 20 0.168 0.8 L-Tryptophan 204 5 0.0245 0.078 L-Tyrosine disodium salt dihydrate 261 29 0.111 0.4 L-Valine 117 20 0.171 0.8 Vitamins Biotin 244 0.2 0.000820 Choline chloride 140 3 0.0214 0.0285 D-Calcium pantothenate 477 0.25 0.000524 0.008 Folic Acid 441 1 0.00227 0.009 i-Inositol 180 35 0.194 .04 Niacinamide 122 1 0.00820 0.033 Para-Aminobenzoic Acid 137 1 0.00730 Pyridoxine hydrochloride 206 1 0.00485 .019 Riboflavin 376 0.2 0.000532 .001 Thiamine hydrochloride 337 1 0.00297 .012 Vitamin B12 1,355 0.005 0.0000037 - Inorganic Salts Calcium nitrate (Ca(NO3)2.4H2O) 236 100 0.424 Magnesium Sulfate (MgSO4) (anhyd.) 120 48.84 0.407 Potassium Chloride (KCl) 75 400 5.33 Sodium Bicarbonate (NaHCO3) 84 2,000 23.81 44 Sodium Chloride (NaCl) 58 6,000 103.45 Sodium Phosphate dibasic (Na2HPO4.7H2O) 268 1,512 5.64 Other Components Glutathione (reduced) 307 1 0.00326 Phenol Red 376.4 5 0.0133 - 10x PBS
Sources of reagents are given in the Materials Section
80 g NaCl
2 g KCl
14.4 g Na2HPO4 anhydrous
2.4 g KH2PO4 anhydrous
Dissolve in 950 ml 18 MOhm water, pH to 7.4, make to 1 L, sterile filter (0.2 μm) - Medium composition for 0.2% 13C6-glucose, 2 mM 12C-Gln (100 ml)
89.2 ml base medium minus tracer (e.g. glucose-free version) (89% concentration of nutrients)
10 ml sterile filtered dialyzed FBS (10% FBS)
0.8 ml 25% sterile filtered 13C6 glucose (0.2 µm) in PBS (10.75 mM glucose final)
1 ml 100x streptomycin/penicillin stock - Medium composition for 0.2% 12C glucose, 2 mM 13C5, 15N2-Gln (100 ml)
88 ml base medium minus tracer (glutamine-free version) (88% concentration of all nutrients)
10 ml sterile filtered dialyzed FBS (10% FBS)
1 ml 0.2 M sterile filtered 13C5, 15N2-glutamine (0.2 µm) in PBS (2 mM final)
1 ml 100x streptomycin/penicillin stock
For hormone sensitive tissues, activated carbon-stripped FBS may be used.
For other concentrations of FBS, adjust the volumes of the FBS and base medium accordingly.
Note: Glutamine stock should be made fresh or stored at -20 °C in small aliquots to avoid repeated freeze and thawing. It forms pyroglutamate on storage in solution even at neutral pH at higher temperatures.
Acknowledgments
This work was supported in part by the following grants: NIH P01 CA163223-01A1, NIH 5R01ES022191-04, NIH 3R01ES022191-04S1, NIH 1U24DK097215-01A1, and the Kentucky Challenge for Excellence. This protocol has been developed based on work described in Xie et al. (2014); Sellers et al. (2015) and Bousamra et al. (2012). The authors declare no conflicts of interest.
References
- Bousamra, M., Day, J., Fan, T. W., Higashi, R. M., Kloecker, G., Lane, A. N. and Miller, D. M. (2012). Clinical aspects of metabolomics. In: The Handbook of Metabolomics. Humana, Vol. 17.
- Fan, T. W. (2012). Considerations of sample preparation for metabolomics investigation. Handbook of Metabolomics 17.
- Fan, T. W. (2010). Metabolomics-edited transcriptomics analysis (meta). In: McQueen, C. A. (ed). Comprehensive Toxicology. Academic Press Vol. 2 685-706.
- Fan, T. W. and Lane, A. N. (2013). Assignment strategies for NMR resonances in metabolomics research. In: Lutz, N., Sweedler, J. V. and Weevers, R. A. (eds). Methodologies for Metabolomics: Experimental Strategies and Techniques. Cambridge University Press.
- Fan, T. W., Lorkiewicz, P. K., Sellers, K., Moseley, H. N., Higashi, R. M. and Lane, A. N. (2012). Stable isotope-resolved metabolomics and applications for drug development. Pharmacol Ther 133(3): 366-391.
- Lane, A. N., Fan, T. W. and Higashi, R. M. (2008). Isotopomer-based metabolomic analysis by NMR and mass spectrometry. Methods Cell Biol 84: 541-588.
- Sellers, K., Fox, M. P., Bousamra, M., 2nd, Slone, S. P., Higashi, R. M., Miller, D. M., Wang, Y., Yan, J., Yuneva, M. O., Deshpande, R., Lane, A. N. and Fan, T. W. (2015). Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. J Clin Invest 125(2): 687-698.
- Warburg, O. (1923). Versuche an überlebendem Carcinomgewebe (Methoden). Biochem Zeitschr 142, 317-333.
- Warburg, O. (1956). On the origin of cancer cells. Science 123(3191): 309-314.
- Xie, H., Hanai, J., Ren, J. G., Kats, L., Burgess, K., Bhargava, P., Signoretti, S., Billiard, J., Duffy, K. J., Grant, A., Wang, X., Lorkiewicz, P. K., Schatzman, S., Bousamra, M., 2nd, Lane, A. N., Higashi, R. M., Fan, T. W., Pandolfi, P. P., Sukhatme, V. P. and Seth, P. (2014). Targeting lactate dehydrogenase--a inhibits tumorigenesis and tumor progression in mouse models of lung cancer and impacts tumor-initiating cells. Cell Metab 19(5): 795-809.
- Zimmermann, M., Sebastian Lange, S., Lampe, J., Smirnow, I., Bitzer, M. and Lauer, U. M. (2012). Precision-cut slices of normal and tumorous liver tissues generated with the Leica VT1200 S vibrating blade microtome. In: Tübingen, M. U. (ed). LeicaBiosystems, Vol. 95.8807 Rev B - Order no. 1495.8807 Tübingen.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Fan, T. W., Lane, A. N. and Higashi, R. M. (2016). Stable Isotope Resolved Metabolomics Studies in ex vivo TIssue Slices. Bio-protocol 6(3): e1730. DOI: 10.21769/BioProtoc.1730.
Category
Cancer Biology > Cellular energetics > Tumor microenvironment > Metabolism
Cancer Biology > Cellular energetics > Animal models > Tissue isolation
Cell Biology > Cell metabolism > Carbohydrate
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link