- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Atomic Force Microscopy (AFM) Analysis of Cell Wall Structural Glycoproteins in vitro
Published: Vol 5, Iss 14, Jul 20, 2015 DOI: 10.21769/BioProtoc.1534 Views: 9501
Reviewed by: Renate WeizbauerFanglian HeAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Live Leaf-Section Imaging for Visualizing Intracellular Chloroplast Movement and Analyzing Cell–Cell Interactions
Yuta Kato [...] Mitsutaka Taniguchi
Aug 5, 2025 2312 Views

Live-Cell Monitoring of Piecemeal Chloroplast Autophagy
Masanori Izumi [...] Shinya Hagihara
Nov 5, 2025 1688 Views

Chloroplast Movement Imaging Under Different Light Regimes With a Hyperspectral Camera
Paweł Hermanowicz [...] Justyna Łabuz
Dec 20, 2025 749 Views
Abstract
Hydroxyproline-rich glycoproteins (HRGPs) are major protein components in dicot primary cell walls and generally account for more than 10% of the wall dry weight. As essential members of the HRGP superfamily, extensins (EXTs) presumably function in the cell wall by assembling into positively charged protein scaffolds (Cannon et al., 2008) that direct the proper deposition of other wall polysaccharides, especially pectins, to ensure correct cell wall assembly (Hall and Cannon, 2002; Lamport et al., 2011a). Extensins are recalcitrant to purification as they are rapidly cross-linked into a covalent network after entering the cell wall but there exists a short time window in which newly synthesized extensin monomers can be extracted (Smith et al., 1984; Smith et al., 1986) by salt elution. A detailed protocol for extraction of extensin and other wall structural proteins has been described earlier (Lamport et al., 2011b). The protocol elaborated here provides an approach to studying the self-assembly of extensins and potentially of other cell wall components in vitro using AFM.
Keywords: Plant cell wallMaterials and Reagents
- Monomeric extensin proteins extracted from different plant cell suspension culture lines. For a detailed extensin extraction protocol see Lamport et al. (2011b).
- Bovine serum albumin (BSA, lyophilized powder, crystallized, purity ≥ 98.0%) (Sigma-Aldrich, catalog number: 05470 )
- Sodium acetate (NaOAc, anhydrous, purity ≥ 99.0%) (Sigma-Aldrich, catalog number: S8750 )
- Acetic acid (HOAc, ACS reagent, purity ≥ 99.7%) (Sigma-Aldrich, catalog number: 320099 )
- Deionized-distilled water (ddH2O)
- 75% ethanol (prepared from 100% ethanol, ACS reagent, purity ≥ 99.5%) (Sigma-Aldrich, catalog number: 459844 )
- Loctite® Epoxy instant mix glue
- 50 mM NaOAc buffer (see Recipes)
- 75% ethanol (see Recipes)
Equipment
- Highly ordered pyrolytic graphite (square shape 5 mm (L) x 5 mm (W) x 1 mm (H) in dimension) (Structure Probe, catalog number: 476HPAB )
- Kimwipes (VWR International, catalog number: 470173498 )
- Plastic petri dishes (100 x 15 mm dimension) (VWR International, catalog number: 25384302 )
- NCS18 silicon probe (75 kHz, 3.5 N/m) (Mikromasch, catalog number: HQ:NSC18/Cr-Au-15)
- Scotch tape (single sided Scotch® MagicTMTape 810)
Note: Available from office supply stores like Staples. - 10 ml sterile syringes (BD Biosciences, catalog number: 309659 )
- 0.2 µm syringe filters (Whatman, catalog number: 6896-2502 )
- 20 ml disposable scintillation glass vials (VWR International, catalog number: 66022065 )
- Eppendorf tubes(1.5 ml) (Eppendorf, catalog number: 022431081 )
- Regular microscope slides (dimension 25 x 75 x 1mm)
- MFP-3D-SA AFM (Asylum Research, model: MFP-3D-SA AFM )
- Stainless steel tweezers (Sigma-Aldrich, catalog number: T5415 )
- Cylinder of nitrogen gas with a pressure regulator and nozzle
- Vortex mixer
- Centrifuge (to fit Eppendorf 1.5 ml tubes)
Software
- IGOR Pro 6 (WaveMetrics, Portland, OR)
Procedure
- Preparations before imaging
- Buffer preparation
- Filter ddH2O through 0.2 µm syringe filter into clean glass vials before use.
- Prepare 50 mM NaOAc buffer (pH 5.2). Filter buffer through 0.2 µm syringe filter into clean glass vials before use.
- Filter ddH2O through 0.2 µm syringe filter into clean glass vials before use.
- Prepare protein stock solutions
Dissolve protein samples to a concentration of 1 mg/ml in filtered buffer as stock solutions using 1.5 ml Eppendorf Protein LoBind Tube (see Note 1). Aliquot stock solutions into 100 µl fractions (see Note 2). Store stock aliquots at -20 °C before use. - Preparation of imaging “substrate” (see Note 3)
Clean one glass slide with ddH2O followed by rinsing with 75% ethanol. Air dry the slide and apply the Loctite® Epoxy instant mix glue to the center of the slide. After a quick mix of the two glue components, carefully remove the HOPG from its case using tweezers and place it on top of the glue. Allow overnight incubation for the tight binding of HOPG to the slide.
- Buffer preparation
- Sample preparation for AFM imaging
- Prepare diluted protein samples
Dilute protein stocks to desired concentration (see Note 4) with corresponding buffer or ddH2O using Eppendorf Protein LoBind Tubes (see Note 1), mix the dilutes by gentle vortexing and then spin down the protein solutions at 3,000 x g for 2 min. - Substrate preparation
HOPG has multiple layers and each layer is atomically flat. Carry out each AFM experiment on a fresh layer. To obtain a new layer of HOPG, simply use Scotch tape to adhere to the previous surface and peel off the old surface. Note that sometimes multiple attempts are required to achieve a visually flat HOPG surface (see Note 5). - Protein binding to HOPG
After obtaining the fresh HOPG surface, place the glass slide with HOPG in a petri dish and deposit 100 µl of protein solution onto the center of the HOPG surface. Cover the petri dish with lid to prevent unnecessary contamination from the surrounding environment. Place moist Kimwipes in the petri dish (avoid direct contact with HOPG) if long incubation time (more than 30 min) is needed to reduce sample evaporation.
Allow the protein solution to incubate on HOPG for one minute or longer (see Note 6) at room temperature. Following incubation, use Kimwipes (see Note 7) to blot away the protein solution and rinse the HOPG surface with 100 µl ddH2O. Blot away the remaining liquid and dry the surface further under a slow steam of N2 gas flow for 2 min. At this point, the sample preparation is complete and the HOPG surface is ready for AFM scanning.
- Prepare diluted protein samples
- AFM imaging of the self-assembly of extensins
Refer to the user’s manual (http://mmrc.caltech.edu/Asylum/Asylum%20MRP-3D%20manual.pdf) from Asylum Research for the operation of the MFP-3D SA AFM. Use “AC mode” (Chapter 6 in the manual, pp 121-142) for the scanning of samples in air. The probe we used was a NCS18 silicon probe from Mikromasch (see Materials and Reagents). For better resolution, Hi'Res-C14 probes (catalog number: Hi'Res-C14/Cr-Au-5) are recommended because of their finer tip radius (~1 nm).
After probe installation, optical detection laser alignment and probe tuning, on the master panel (see pp136 in the manual), select the initial scan parameters as follows:
Scan Size = 5.00 µm
Scan Rate = 0.75 Hz (see Note 8)
Scan Points/Lines = 256
Set Point = 800.00 mV
Integral Gain = 4 (see Note 9)
Engage the AFM probe to sample surface by lowering the AFM head and start scanning. Here note that the “Drive Amplitude/Frequency” values on the master panel are the values automatically filled in by the instrument upon the finishing of probe tuning, thus no changes of these values are needed.
After the start of scanning, first allow the system to scan for 2 min to stabilize the probe and adjust the instrument to surrounding environmental vibrations. Then manually lower the “Set Point” value until surface features start to appear on the height image. Imaging optimization can be achieved by adjusting “Set Point” and “Integral Gain” values. Finally, the “Scan Points” and “Scan Lines” can be increased to 512 or even 1,024 (see Note 10) for more pixels in each image thus enhance the image quality.
For the determination of self-assembly pattern, take at least five images at five different locations on the HOPG surface for each sample. - Image processing
Images need to be flattened using the “flatten” function in IGOR Pro software before any measurements. Information of height (i.e. the diameter) and length of a molecule can be directly measured on the images using the IGOR Pro software. The default color setting of the images is grey but IGOR Pro has a variety of built-in color schemes that can be used to false-color images afterwards. In the meantime, the contrast and brightness of each image can also be adjusted for a better presentation of the image.
Representative data
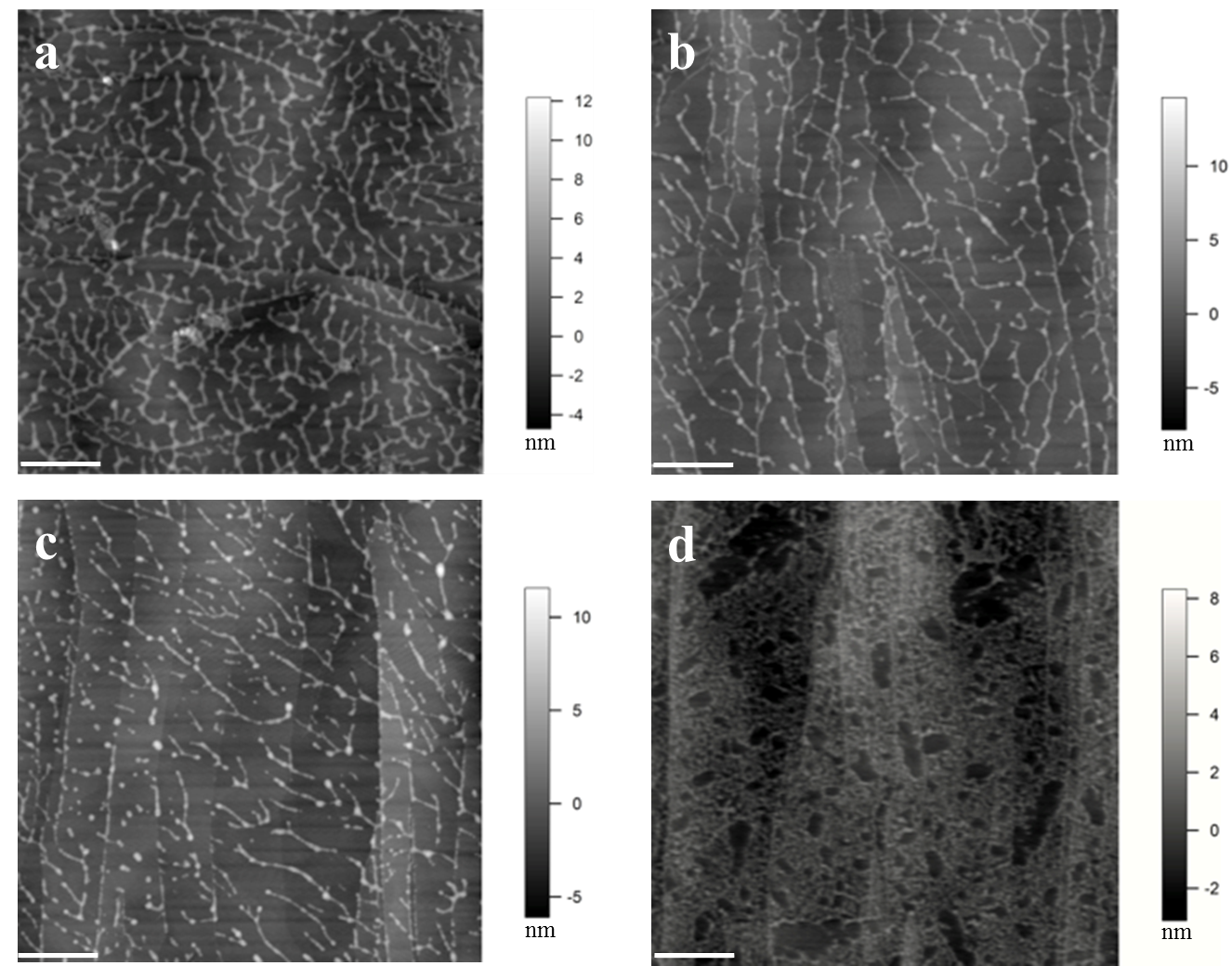
Figure 1. Images of extensin self-assembly. (a) Monomeric extensin precursor 1 from tomato suspension culture (TOMP1). (b) Extensin analog YK8. (c) Extensin analog FK9. (d) BSA used as a control. YK8 and FK9 are extensin analogs that contain 8 repeats of SOOOOSOSOOOOYYYK and 9 repeats of SOOOOSOSOOOOFFFK in their protein sequences, respectively, and are purified from tobacco BY2 cells in culture (Held et al., 2004). One letter amino acids codes: Tyrosine (Y), Lysine (K), Phenylalanine (F), Serine (S), Hydroxyproline (O). All proteins were imaged at 10 μg/ml in pH 5.2 NaOAc buffer, following deposition for 1 min. The white scale bar: 500 nm. TOMP1 and the extensin analogs all showed a head to tail dendritic self-assembly similar to that of EXTENSIN 3 from Arabidopsis (Cannon et al., 2008), while BSA showed no such assembly but only protein aggregation. Red arrow in image (d) indicates the scattered single sphere shaped BSA molecules.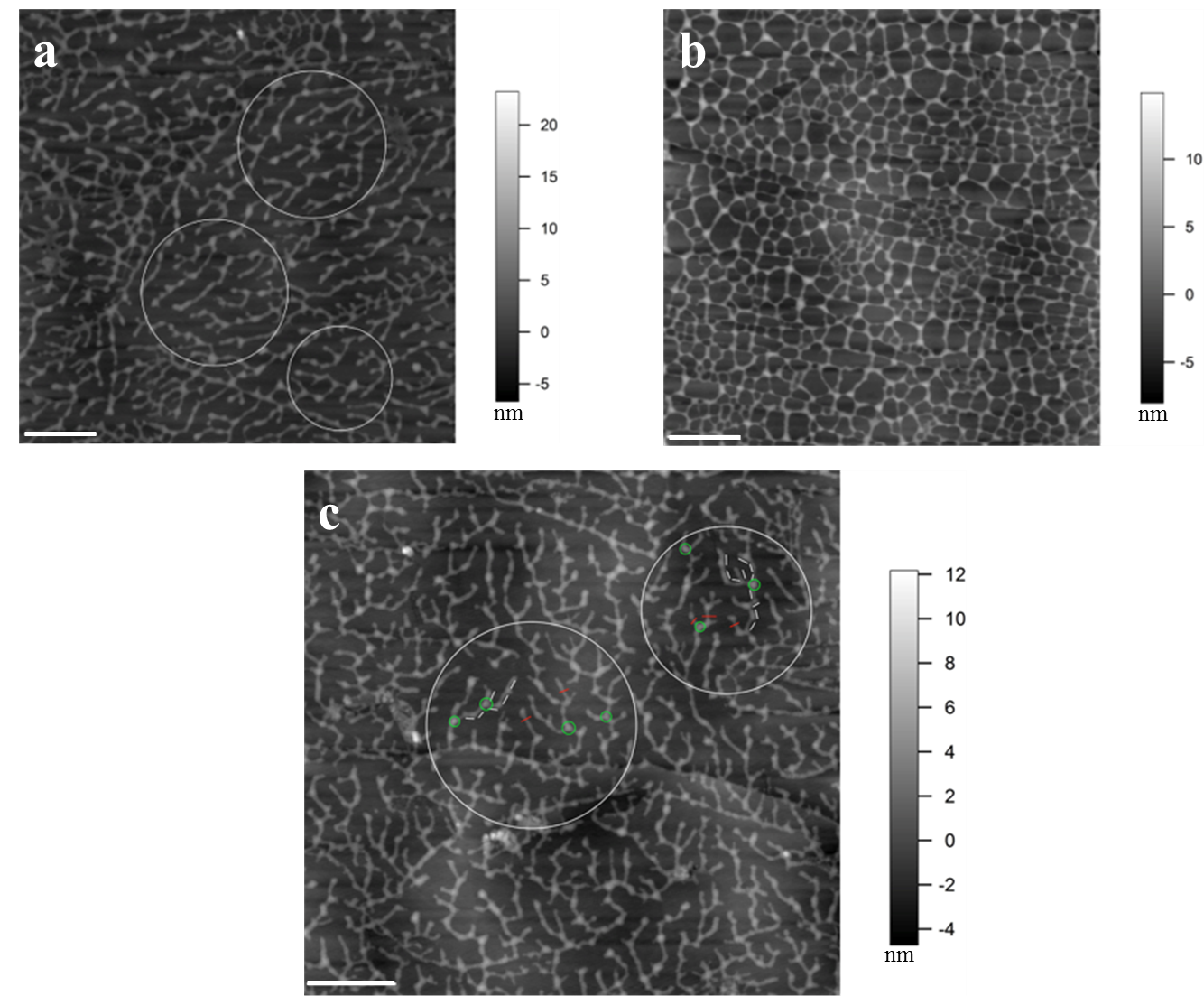
Figure 2. Concentration and deposition time dependent self-assembly of TOMP1 in pH 5.2 NaOAc buffer. a. TOMP1 showed head to tail dendritic self-assembly after 10 min deposition on HOPG at 5 µg/ml. b. At a higher concentration (10 µg/ml) TOMP1 assembled into a porous network in the same time period, indicating the extent of self-assembly is concentration dependent. c. Similar assembly of TOMP1 as in (a), highlighted by the white circles, was observed when 10 µg/ml of TOMP1 was deposited for 1 min, indicating that the extent of TOMP1 self-assembly also followed a time dependent manner. Examples of measurements for TOMP1 self-assembly: (1) The average segment length, highlighted by white bars in the circles in image (c), is 78.2 ± 4.2 nm this is in agreement with previously electron microscopy measurements of the TOMP1 polypeptide length at about 79 nm (Heckman et al., 1988). This observation indicates one TOMP1 molecule occurs in each segment. (2) The average single molecule height, highlighted by red bars in image (c), is 2.7 ± 0.2 nm. This value corresponds to the diameter of single TOMP1 molecule. (3) The average height of the connecting points for three molecules, highlighted by green circles in image (c), is 5.2 ± 0.2 nm. This value indicates that the TOMP1 molecules form lateral and overlapping associations up to two molecules deep. The white scale bar: 500 nm
Notes
- The protein concentrations used in stock and imaging are relatively low. The use of Eppendorf Protein LoBind tubes will prevent non-specific binding of protein to the tubes thus resulting in a more accurate protein amount used in experiments.
- Repeated freezing-thawing will lead to protein degradation and unexpected aggregation. Use one aliquot per experiment to help prevent such damage to the protein stocks.
- In AFM imaging, the surface on which sample solutions are deposited is called “substrate”. We used HOPG for imaging of proteins due to its hydrophobicity, which allows the stable binding of proteins to the surface. To study other wall components, for instance wall polysaccharides and cell wall itself, other substrates such as mica (for polysaccharides) or charged glass slides (for cell wall) can be used as alternatives to ensure the binding of sample to the substrate surface.
- Depending on the purpose of the experiments, various protein concentrations can be used. From our experience, a low protein concentration (5 or 10 µg/ml) will benefit in the observation of extensin single molecular assembly while a higher concentration (50 µg/ml) will result in an orderly formed network.
- HOPG will get thinner after multiple uses, thus making it progressively more difficult to obtain a new visually flat surface. Based on its original height dimension (1 mm), a replacement is recommended when the HOPG reaches less than 0.4 mm in height.
- Similar to Note 4, the length of incubation time also determines the pattern of observed self-assembly. Usually after longer incubation, a well-formed extensin network is observed. From our experience, the attachment of extensin molecules to the HOPG surface happens quickly. Incubation time as short as 1 min is sufficient for the binding of extensins to HOPG.
- The use of regular paper towels is not recommended since there might be unexpected fibers or debris that might contaminate the sample surface.
- Use a slow scan rate at first to protect the probe, the “Scan Speed” is automatically set upon the selection of “Scan Rate”.
- Use a low “Integral Gain” in the beginning to protect the probe. This value can be elevated in a later scan for better image quality.
- “Scan Points” and “Scan Lines” determine the number of pixel point on each line and the number of lines scanned in the image, in most of the cases these two values should be kept the same.
Recipes
- 50 mM NaOAc buffer
0.41 g NaOAc
100 ml distilled water
Adjust pH to 5.2 with HOAc - 75% ethanol
Add 25 ml water to 75 ml 100% ethanol to make 100 ml 75% ethanol
Acknowledgments
This protocol was adapted with modification from Cannon et al. (2008). Funding of this work was from National Science Foundation (#IOS955569 to M.J.K and IOS0955805 to M.C.C).
References
- Cannon, M. C., Terneus, K., Hall, Q., Tan, L., Wang, Y., Wegenhart, B. L., Chen, L., Lamport, D. T., Chen, Y. and Kieliszewski, M. J. (2008). Self-assembly of the plant cell wall requires an extensin scaffold. Proc Natl Acad Sci USA 105(6): 2226-2231.
- Hall, Q. and Cannon, M. C. (2002). The cell wall hydroxyproline-rich glycoprotein RSH is essential for normal embryo development in Arabidopsis. Plant Cell 14(5): 1161-1172.
- Heckman, J. W., Terhune, B. T. and Lamport, D. T. (1988). Characterization of native and modified extensin monomers and oligomers by electron microscopy and gel filtration. Plant Physiol 86(3): 848-856.
- Held, M. A., Tan, L., Kamyab, A., Hare, M., Shpak, E. and Kieliszewski, M. J. (2004). Di-isodityrosine is the intermolecular cross-link of isodityrosine-rich extensin analogs cross-linked in vitro. J Biol Chem 279(53): 55474-55482.
- Lamport, D. T., Kieliszewski, M. J., Chen, Y. and Cannon, M. C. (2011a). Role of the extensin superfamily in primary cell wall architecture. Plant Physiol 156(1): 11-19.
- Lamport, D. T., Tan, L. and Kieliszewski, M. J. (2011b). Structural proteins of the primary cell wall: extraction, purification, and analysis. Methods Mol Biol 715: 209-219.
- Smith, J. J., Muldoon, E. P. and Lamport, D. T. A. (1984). Isolation of extensin precursors by direct elution of intact tomato cell suspension cultures. Phytochem 23, 1233-1239.
- Smith, J. J., Muldoon, E. P., Willard, J. J. et al. (1986). Tomato extensin precursors P1 and P2 are highly periodic structures. Phytochem 5, 1021-1030.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Chen, Y., Chen, L., Kieliszewsk, M. J. and Cannon, M. C. (2015). Atomic Force Microscopy (AFM) Analysis of Cell Wall Structural Glycoproteins in vitro. Bio-protocol 5(14): e1534. DOI: 10.21769/BioProtoc.1534.
Category
Plant Science > Plant cell biology > Cell imaging
Plant Science > Plant cell biology > Cell structure
Cell Biology > Cell imaging > Atomic force microscopy
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










