- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Large-scale Phenotypic Profiling of Gene Deletion Mutants in Candida glabrata
(*contributed equally to this work) Published: Vol 5, Iss 14, Jul 20, 2015 DOI: 10.21769/BioProtoc.1530 Views: 10791
Reviewed by: Sadri ZnaidiFanglian He

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In vitro Cell Wall Stress Assay for Fusarium oxysporum
Elena Pérez-Nadales and Antonio Di Pietro
Sep 5, 2016 10885 Views

Single-step Precision Genome Editing in Yeast Using CRISPR-Cas9
Azat Akhmetov [...] Edward M Marcotte
Mar 20, 2018 20389 Views

Competition Assays to Quantify the Effect of Biocontrol Yeasts against Plant Pathogenic Fungi on Fruits
Electine Magoye [...] Florian M. Freimoser
Feb 5, 2020 5418 Views
Abstract
Here, we describe a method enabling the phenotypic profiling of genome-scale deletion collections of fungal mutants to detect phenotypes for various stress conditions. These stress conditions include among many others antifungal drug susceptibility, temperature-induced and osmotic as well as heavy metal or oxidative stress. The protocol was extensively used to phenotype a collection of gene deletion mutants in the human fungal pathogen Candida glabrata (C. glabrata) (Schwarzmüller et al., 2014).
Keywords: PhenotypingMaterials and Reagents
- C. glabrata strain (ATCC2001, clinical isolates)
- Sterile water (double distilled)
- Bacto™ peptone (BD Biosciences, catalog number: 211820 )
- Bacto™ yeast extract (BD Biosciences, catalog number: 212720 )
- Bacto™ agar (BD Biosciences, catalog number: 214030 )
- Difco™ yeast nitrogen base (YNB) (BD Biosciences, catalog number: 233520 )
- Glucose (Merck KGaA, catalog number: 108337 )
- Adenine (Sigma-Aldrich, catalog number: A8626 )
- L-arginine (Sigma-Aldrich, catalog number: A5006 )
- L-tyrosine (Sigma-Aldrich, catalog number: T3754 )
- L-isoleucine (Sigma-Aldrich, catalog number: I2752 )
- L-phenylalanine (Sigma-Aldrich, catalog number: P2126 )
- L-glutamic acid(Sigma-Aldrich, catalog number: G1251 )
- L-aspartic acid (Sigma-Aldrich, catalog number: A9256 )
- L-threonine (Sigma-Aldrich, catalog number: T8625 )
- L-serine (Sigma-Aldrich, catalog number: S4500 )
- L-valine (Sigma-Aldrich, catalog number: V0500 )
- L-methionine (Sigma-Aldrich, catalog number: M9625 )
- Glycerol (Sigma-Aldrich, catalog number: G5516 )
- YPD media (see Recipes)
- Solid YPD media (see Recipes)
- 2x YPD media (see Recipes)
- SC media (see Recipes)
- Solid SC media (see Recipes)
- Amino acid mix (see Recipes)
- 15% glycerol solution (see Recipes)
Equipment
- Square plates (PlusPlate) (40 ml/plate) (Singer Instruments, catalog number: PLU-001 )
- Pin Pad (RePad 96 Long) (Singer Instruments, catalog number: REP-001 )
- 96-well plates (200 µl/well) (Starlab, catalog number: CC76827596 )
- Petri dishes (92 mm) (SARSTEDT AG, catalog number: 82.1473 )
- Culture tubes (Starlab, catalog number: I14850810 )
- Yeast replica robot (Singer Instruments, catalog number: RoToR HDA )
- Plate reader (Victor3V) (PerkinElmer, catalog number: 1420040 )
- 48-well replica stamp (V&P Scientific, catalog number: VP 408H )
- 96-well replica stamp (V&P Scientific, catalog number: VP 407 )
- Library copier (guide for stamps) (V&P Scientific, model: VP 381 )
- Plate mixer (Eppendorf, catalog number: 5353000014 )
- Rotary shaker for culture flasks (innova44) (New Brunswick Scientific, catalog number: M1282-0002 )
- Incubator (Heraeus Instruments, catalog number: B6120 )
Procedure
- Arraying and storage
- For arraying deletion strains in the 96-well format see the Note section. Arrayed strains can be suspended in 15% glycerol and stored frozen at minus 70 °C in 96-well plates.
- Frozen stocks can be either handled by manual replicating tools or by an automated system such as the RoToR HDA by Singer Instruments. The latter offers a higher reproducibility and reduces the risk of contamination when compared to manual systems. Therefore, this protocol for large-scale screening was optimized for the work with the Singer HDA RoToR replica-plating robot.
- For arraying deletion strains in the 96-well format see the Note section. Arrayed strains can be suspended in 15% glycerol and stored frozen at minus 70 °C in 96-well plates.
- Initial large-scale screening (on solid YPD media)
- For initial pilot screenings, prepare square plates containing the screening compounds and control plates lacking compounds as shown in Figure 1. The concentration of the compound for the solid screening plate is usually twice the concentration of the IC50. However, the exact concentration should be determined prior to the large-scale screening by plating control strains.
- To exclude replica-plating errors, at least two plates should be prepared for each condition (stress-inducing compound, stress-inducing media, temperature, etc.).
- Source plates can be either liquid media (96-well microtiter plates) or solid media (square plates).
- To exclude replica-plating errors, at least two plates should be prepared for each condition (stress-inducing compound, stress-inducing media, temperature, etc.).
- Dry plates on a clean bench for 10 min, for light-sensitive compounds turn off lights.
- Make sure that no droplets of condensed water are on the plates or lids before using them.
- Label plates before spotting strains.
When using a Singer robot use the following settings in the ROTOR HDA software:
- For solid source plates activate the “offset” function, to avoid pins taking cells from exactly the same spot.
- If liquid source plates are used, enable the “wet mix” option in the software menu (diameter: 1 mm; speed: 3 mm/sec; cycles: 1; Travel: 2D). Alternatively, liquid source plates can be shaken on a MixMate for 15 sec at 750 rpm.
- If the so-called RePads are used for more than one target plate (“Recycle Mode”), the “Revisit Source” option should be enabled as well.
- Make sure that no droplets of condensed water are on the plates or lids before using them.
- After plating, turn plates upside down as soon as spots have dried. In case of solid source plates, the target plates can be turned right away. Incubate plates for 72 h at 30 °C without shaking and inspect colony growth. Pictures should be taken of every plate at 24, 48 and 72 h. Visually compare the growth of each mutant strains and controls under stress and without stress with the growth of wild type strain(s). Important points that should be considered are shown in Figures 2 and 3.
- For initial pilot screenings, prepare square plates containing the screening compounds and control plates lacking compounds as shown in Figure 1. The concentration of the compound for the solid screening plate is usually twice the concentration of the IC50. However, the exact concentration should be determined prior to the large-scale screening by plating control strains.
- Serial dilution spot assays (solid SC media)
- For refined screening, prepare at least two SC media plates for each condition. Prior to their use, dry them on a clean bench. Grow overnight cultures of sensitive and wild type strains from the 96-well screening in SC medium at 30 °C with shaking at 200 rpm (200 rpm are used for growth in culture tubes; a total amount of 5 ml for each culture is sufficient). Dilute cultures into fresh SC medium resulting in an OD600 of ≈ 0.2 and regrow them to an OD600 of ≈ 1.0. Prepare 1:10 serial dilutions of deletions strains in sterile water in a 96-well plate, starting with an OD600 of 0.1.
Table 1. Serial dilution of cells
OD600
Dilution
Cells/m
0.1
1:1
3 x 106
0.01
1:10
3 x 105
0.001
1:100
3 x 104
0.0001
1:1,000
3 x 103
- Spotting uses a 48-well metal pin replicator, also referred to as “frogger” or “hopper”.
- Sterilize the tool by submerging the pins in EtOH and brief flaming over a bunsen burner. Repeat this process three times and let the pins cool down.
- Stir the serial dilution in the 96-well plate using the replicator and retract the pins from the wells without touching the wall of the wells.
- A droplet of cell suspension will remain on each tip of the replicator.
- Place the replicator on a fresh screening plate and apply gentle pressure. Lift the replicator straight up, avoid an angle and repeat the process for other plates.
- Incubate the plates as described above at step B3 and takes pictures of every plate after 24, 48 and 72 h of incubation.
- Sterilize the tool by submerging the pins in EtOH and brief flaming over a bunsen burner. Repeat this process three times and let the pins cool down.
- Alternatively, spotting can be performed using a multichannel or a single-channel pipette. When using a pipette, a grid should be placed under the plate to make sure the drops are placed in a straight line.
- After the drops have dried, turn the plates upside down and incubate them at 30 °C for 72 h without shaking. Compare growth of the serial dilutions of each mutant and the wild type control under stress and without stress.
- For refined screening, prepare at least two SC media plates for each condition. Prior to their use, dry them on a clean bench. Grow overnight cultures of sensitive and wild type strains from the 96-well screening in SC medium at 30 °C with shaking at 200 rpm (200 rpm are used for growth in culture tubes; a total amount of 5 ml for each culture is sufficient). Dilute cultures into fresh SC medium resulting in an OD600 of ≈ 0.2 and regrow them to an OD600 of ≈ 1.0. Prepare 1:10 serial dilutions of deletions strains in sterile water in a 96-well plate, starting with an OD600 of 0.1.
- Microdilution of compounds (liquid media)
- Grow overnight cultures of deletion and control wild type strains in YPD at 30 °C with shaking at 200 rpm (as described in step C1, 200 rpm are used for culture tubes; 5 ml of each culture are sufficient). Dilute strains in fresh YPD media resulting in an OD600 of ≈ 0.2 and regrow them until an OD600 of ≈ 1.0. Dilute cell suspensions in 2x YPD to a final concentration of 2.5 x 103 cells/ml.
- Prepare a series of ten two-fold dilutions of the compound to be tested in sterile water. Fill each well with 100 µl of compound dilution or water. Rows 11 and 12 should only contain sterile water and be used as growth and no-growth control.
- Add 100 µl cell suspension in 2x YPD to the wells of the 96-well plate. Seal the plate using a clear breathable foil and incubate the plate at 30 °C without shaking for 48 h. Measure the OD600 on a plate reader after 24 and 48 h. Before reading the plate mix it at 1,000 rpm for 15 sec. Calculate compound concentrations required to obtain 50 % growth inhibition (IC50) for each strain.
- Grow overnight cultures of deletion and control wild type strains in YPD at 30 °C with shaking at 200 rpm (as described in step C1, 200 rpm are used for culture tubes; 5 ml of each culture are sufficient). Dilute strains in fresh YPD media resulting in an OD600 of ≈ 0.2 and regrow them until an OD600 of ≈ 1.0. Dilute cell suspensions in 2x YPD to a final concentration of 2.5 x 103 cells/ml.
- Evaluation of results
- Since the large-scale screening may result in false-positive hits, it needs independent unbiased inspection of primary results. Therefore, the screenings described in sections C-D should be independently analyzed by at least two persons.
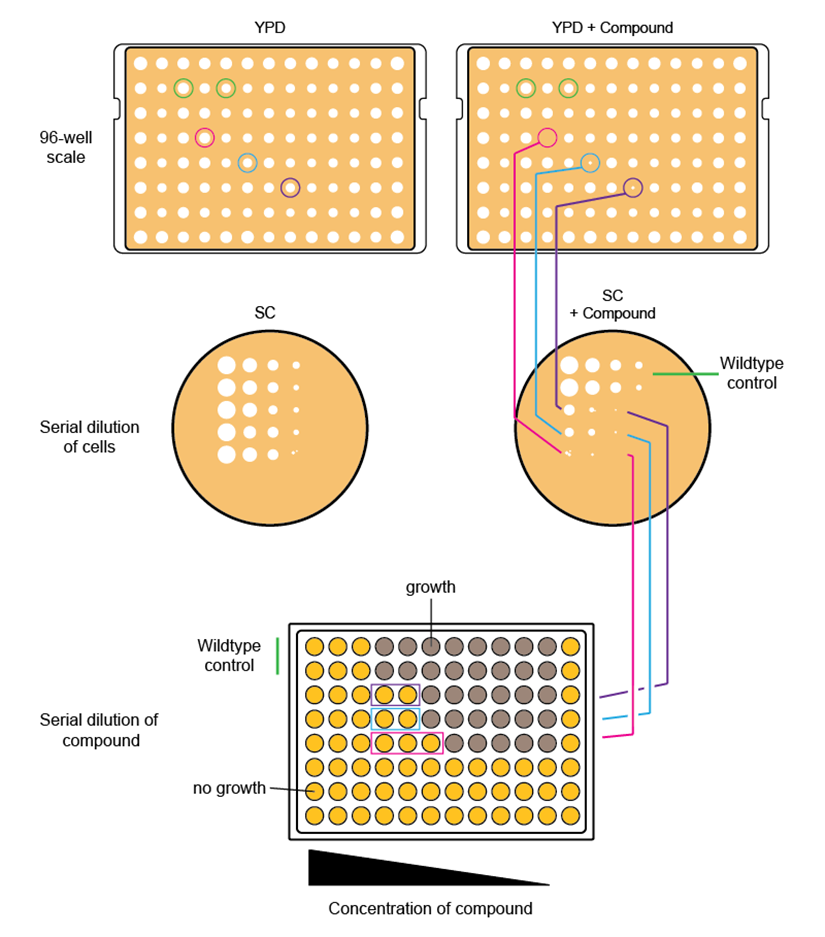
Figure 1. Screening process-from large scale to single strains. Putative hits (purple, blue, pink) are identified by comparing growth of strains on a control plate (YPD only) with a plate supplemented with screening compounds. In addition, deletion strains must be compared to the corresponding wild types (first two rows). These strains are then used for a refined screening in liquid assays through serial dilution on solid media to confirm primary phenotypes.
- To validate the link between a genotype and a phenotype more experiments are necessary. These experiments include the complementation of deletion strains and the deletion of a single gene in different backgrounds.
- Since the large-scale screening may result in false-positive hits, it needs independent unbiased inspection of primary results. Therefore, the screenings described in sections C-D should be independently analyzed by at least two persons.
Notes
- Plate layouts
When designing the layout several aspects should be considered.
Outer spots grow faster.
Outer spots are framed red in Figure 2 and show a faster growth when compared to spots in inner rows and columns.
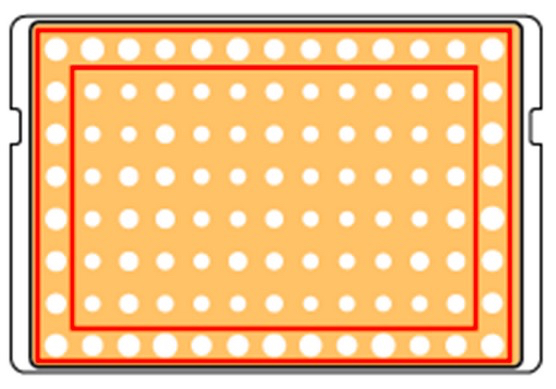
Figure 2. Faster growth of outer spots
This phenomenon may hamper comparison of growth phenotypes of deletion strains to the growth of a wild type strain. In the worst case, this may lead to missing relevant phenotype. This problem can be solved by filling all outer rows / columns with a dummy strain (e.g. the wild type). Therefore, all strains that are to be screened are surrounded on all sides and experience the same growth conditions. Of note, this procedure yields only 60 usable spots. Alternatively, a 96-well plate can be spotted in quadruplicate for each deletion in the 384-well format using the Singer HDA RoToR. This method will result in each deletion strain being spotted 4 times on the agar plate as illustrated in Figure 3.
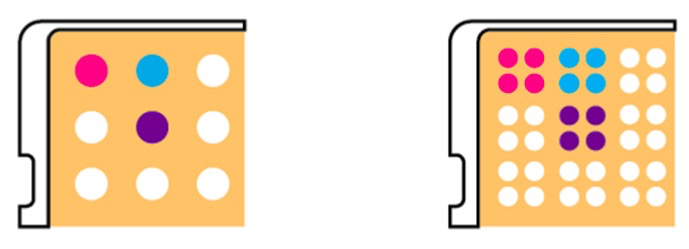
Figure 3. 96-well to 384-well format. The 384-well format (right panel) offers two distinct advantages when compared to the 96-well format (left panel). Only the outer spots grow faster, so every original spot has at least one spot now that grows normally. In addition, it can be easier to identify growth phenotypes due to the uniform appearance of growth patterns.
Include empty spots for controls
To compare results, at least the background strain should be spotted on each plate. Since this strain is usually derived from a wild type strain, the wild type should also be included. Moreover, it is very useful to include empty spots on each plate and to add additional positive and negative control strains for a specific screening. For instance, it can be very helpful to temporarily add a known hypersensitive strain for a specific screening as positive control [for an azole screening a pdr1∆ (CAGL0A00451g) strain is a great control].
- Liquid source plates are preferable to solid source plates, since the reproducibility is higher when using liquid sources plates. We experienced that the amount of cells can be inconsistent when using solid source plates. Liquid source plates can be shaken prior to plating, resulting in a very homogenous source cell suspension.
- There are various standardized protocols available for the microdilution assays to quantify drug susceptibilities including those of the Clinical and Laboratory Standards Institute (Fothergill et al., 2011) or the European Committee on Antimicrobial Susceptibility Testing (Arendrup et al., 2012).
- A video of the Singer RoToR in use is available at:
https://www.youtube.com/watch?v=1JabqneqXPE.
- Stocks for water insoluble compounds can be prepared in the appropriate solvent (e.g. DMSO). However, the dilution should be done in water. Otherwise, the wells would contain up to 50% of solvent.
Recipes
- YPD media (yeast extract peptone dextrose)
25 g/LBacto™ peptone
12.5 g/LBacto™ yeast extract
2%glucose
- Solid YPD media
25 g/LBacto™ peptone
12.5 g/LBacto™ yeast extract
2%glucose
2%agarose
- 2x YPD media
50 g/LBacto™ peptone
25 g/LBacto™ yeast extract
4%glucose
- SC media (synthetic complete)
3.4 g/LDifco™ YNB
2.9 g/Lamino acid mix
2 %glucose
- Solid SC media
3.4 g/LDifco™ YNB
2.9 g/Lamino acid mix
2 %glucose
2 %agarose
- Amino acid mix (g per 11.7 g total mix)
0.4 gadenine
0.2 gL-arginine
0.3 gL-tyrosine
0.3 gL-isoleucine
0.5 gL-phenylalanine
1.0 gL-glutamic acid
1.0 gL-aspartic acid
2.0 gL-threonine
4.0 gL-serine
1.5 gL-valine
0.5gL-methionine
- 15% glycerol solution
150 ml/L glycerol
850 ml/L water
Acknowledgments
This work was supported by the Austrian Science Foundation FWF through the ERA-Net Pathogenomics project FunPath (FWF-API-0125), and in part by grants from the Christian Doppler Society, the FP7 EC project FUNGITECT, the Marie-Curie ITN ImResFun (MC-ITN-606786) and the FWF Project FWF-P25333 "Chromatin" to KK.
References
- Arendrup, M. C., Cuenca-Estrella, M., Lass-Florl, C., Hope, W. and Eucast, A. (2012). EUCAST technical note on the EUCAST definitive document EDef 7.2: method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for yeasts EDef 7.2 (EUCAST-AFST). Clin Microbiol Infect 18(7): E246-247.
- Fothergill, A. W. (2011). Antifungal susceptibility testing: clinical laboratory and standards institute (CLSI) methods. Interactions of Yeasts, Moulds, and Antifungal Agents 65-74.
- Schwarzmuller, T., Ma, B., Hiller, E., Istel, F., Tscherner, M., Brunke, S., Ames, L., Firon, A., Green, B., Cabral, V., Marcet-Houben, M., Jacobsen, I. D., Quintin, J., Seider, K., Frohner, I., Glaser, W., Jungwirth, H., Bachellier-Bassi, S., Chauvel, M., Zeidler, U., Ferrandon, D., Gabaldon, T., Hube, B., d'Enfert, C., Rupp, S., Cormack, B., Haynes, K. and Kuchler, K. (2014). Systematic phenotyping of a large-scale Candida glabrata deletion collection reveals novel antifungal tolerance genes. PLoS Pathog 10(6): e1004211.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Istel, F., Schwarzmüller, T., Tscherner, M. and Kuchler, K. (2015). Large-scale Phenotypic Profiling of Gene Deletion Mutants in Candida glabrata. Bio-protocol 5(14): e1530. DOI: 10.21769/BioProtoc.1530.
Category
Microbiology > Antimicrobial assay > Antifungal assay
Systems Biology > Genomics > Functional genomics
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link