- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Pyrosequencing Approach for SNP Genotyping in Plants Using a M13 Biotinylated Primer
Published: Vol 5, Iss 10, May 20, 2015 DOI: 10.21769/BioProtoc.1473 Views: 10529
Reviewed by: Fanglian HeKabin XieKanika Gera

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Experimental Pipeline for SNP and SSR Discovery and Genotyping Analysis of Mango (Mangifera indica L.)
Michal Sharabi-Schwager [...] Ron Ophir
Aug 20, 2016 11552 Views

Investigation of Transposon DNA Methylation and Copy Number Variation in Plants Using Southern Hybridisation
Vivek Hari Sundar G. and P. V. Shivaprasad
Jun 5, 2022 3404 Views

CAPS-Based SNP Genotyping for Nitrogen-Response Phenotypes in Maize Hybrids
Jannis Jacobs [...] Peter K. Lundquist
Dec 20, 2025 543 Views
Abstract
Single Nucleotide Polymorphisms (SNPs), which constitute single base-pair variations in the DNA sequence, are the most abundant molecular markers in plant and animal genomes. They are becoming the markers of choice for genotyping in all fields of molecular biology, as they are easily prone to automation and high throughput, for example through pyrosequencing. This technology is accurate, flexible and can be easily automated. However, the need for primers labelled with biotin, promptly rise the cost of any methodology employing a pyrosequencing approach. In this protocol we described an improved, efficient, reliable and cost-effective pyrosequencing protocol, based on a universal M13 biotinylated primer, for SNP genotyping in plants.
Keywords: SNPMaterials and Reagents
- DNA (25 ng/µl) e.g. from two-weeks old barley seedlings; or according to PCR practice of each particular organism
- PCR reagents
- 0.5 U Taq DNA polymerase (Solis Biodyne FIREPol®, catalog number: 01-01-01000 ) or HotStart Taq polymerase (Solis Biodyne HOT FIREPol®, catalog number: 01-02-01000 ; any other suppliers should be also satisfactory) with the corresponding 10x PCR buffer (supplied with Taq DNA polymerase)
- 25 mM MgCl2 (supplied with Taq DNA polymerase)
- dNTPs (10 µM each) (Thermo Fisher Scientific, catalog number: R0182 )
- Forward specific primer tailed at the 5´end with a universal M13 tail (5´- CACGACGTTGTAAAACGAC-3´) (desalted) (1 µM)
- Reverse specific primer (desalted) (10 µM)
- Biotinylated universal primer with a complementary sequence to the M13 tail (10 µM) (Metabion, Planegg/Steinkirchen)
- 0.5 U Taq DNA polymerase (Solis Biodyne FIREPol®, catalog number: 01-01-01000 ) or HotStart Taq polymerase (Solis Biodyne HOT FIREPol®, catalog number: 01-02-01000 ; any other suppliers should be also satisfactory) with the corresponding 10x PCR buffer (supplied with Taq DNA polymerase)
- Agarose (Sigma-Aldrich, catalog number: A9539 )
- Ethidium bromide (Roche Diagnostics, catalog number: HP46.2 )
- Pyrosequencing reagents
- Sequencing primer (desalted) (10 µM)
- Streptavidin Sepharose HP (GE Healthcare, catalog number: 17-5113-01 )
- Annealing buffer (QIAGEN, catalog number: 979009 ) (see Recipes)
- Binding buffer (QIAGEN, catalog number: 979006 ) (see Recipes)
- Denaturation solution (QIAGEN, catalog number: 979007 ) (see Recipes)
- Washing buffer (QIAGEN, catalog number: 979008 ) (see Recipes)
- High-purity water (Milli-Q)
- 70% ethanol
- PyroMark Gold Q96 Reagents (enzyme mixture, substrate mixture and nucleotides) (QIAGEN, catalog number: 972807 )
- Sequencing primer (desalted) (10 µM)
- Gel running buffer (see Recipes)
- 6x DNA loading buffer (see Recipes)
Equipment
- Thermal Cycler (Applied Biosystems)
- Horizontal Electrophoresis system (Bio-Rad Laboratories)
- PyroMark Q96 instrument (QIAGEN, catalog number: 9001525 )
- PyroMark Q96 plate (QIAGEN, catalog number: 979002 )
- PyroMark Q96 Vacuum Workstation (220 V) (QIAGEN, catalog number: 9001529 )
- Vacuum pump (KNF Lab, Typ N816.1.2 KN.18)
- Orbital shaker for microtiter plates
- Heating block (80 °C)
- PyroMark Q96 Cartridge (QIAGEN, catalog number: 979004 )
- PCR 96-well plate (Greiner Bio-One, catalog number: 652250 )
Software
- PyroMark Assay Design Software V.1.0.6. (QIAGEN)
Procedure
- Use the PSQ Assay Design Software for the design of forward, reverse and sequencing primers.
- Enter or import the sequence of interest into the “Sequence editor” with the SNP positions represented by IUPAC codes or the following format: C/T.
- The amplicon size should be between 150 and 200 bp.
- The tailed primer (forward or reverse) should not be longer than 20 bp.
- The sequencing primer should be designed between one and five nucleotides of the target SNP.
- Select those primers with the highest scores assigned by the software according to their quality for PCR and Pyrosequencing.
- Export the primers sequence to an excel file and manually add the universal tail (5´-CACGACGTTGTAAAACGAC-3´) to the 5´end of the forward or reverse primer.
- Copy and paste into the excel file the “sequence to analyze”, which will be required later during the run by the PSQ 96 instrument in order to establish the dispensation order for the different nucleotides.
- Enter or import the sequence of interest into the “Sequence editor” with the SNP positions represented by IUPAC codes or the following format: C/T.
- Prepare a PCR mix in a final volume of 25 µl: 1x PCR buffer, 2.5 mM MgCl2, 0.2 mM dNTPs, 0.02 µM of tailed-forward primer, 0.2 µM of reverse primer, 0.18 µM of biotinylated M13 primer, 0.5 U of Taq polymerase and 50 ng of DNA. Even if other DNA source is considered (e.g. PCR product), a minimum of 50 ng should be used as well.
- Run the following PCR program: 5 min at 94 °C; 12 cycles with 30 sec at 94 °C, 30 sec at 62 °C (touchdown of 0.5 °C/cycle for initial 12 cycles - final annealing of 56 °C for remaining 35 cycles), 30 sec at 72 °C; and a final extension step of 10 min at 72 °C. At this step, biotin is being incorporated into the PCR product, as described by Schuelke (2000) for fluorescent labelling of PCR fragments.
- Run 3 µl of the PCR product in an agarose gel (1.5%) stained with ethidium bromide.
- Check the presence of a single and strong DNA fragment with the expected size. If there is any unspecific product, which is highly unusual if the primer design was performed accurately, a HotStart Taq polymerase might be used with the same PCR mix described above, except for MgCl2 (2 mM), dNTPs (0.125 mM) and Taq (0.6 U). The PCR program in this case is as follows: 15 min at 95 °C; 12 cycles with 30 sec at 95 °C, 30 sec at 62 °C (touchdown of 0.5 °C/cycle for initial 12 cycles - final annealing of 56 °C for remaining 45 cycles), 30 sec at 72 °C; and a final extension step of 10 min at 72 °C.
- Before starting the pyrosequencing assay, let the buffers reach room temperature.
- Add high-purity water to the PCR samples to a final volume of 40 µl.
- Prepare a mixture of Streptavidin Sepharose beads (3 µl per sample) and binding buffer (37 µl per sample).
- Add 40 µl of the mixture prepared in step 8 to each well of the PCR plate.
- Seal the plate and shake the PCR plate constantly at ca. 1,000 rpm for 10 min at room temperature.
- Add 40 μl of 0.4 μM sequencing primer in annealing buffer to each well of the PSQ plate that is to be used.
- After shaking, place the PCR plate and PSQ plate on the vacuum work station.
- Fill each of the five separate troughs in the vacuum workstation with the following solutions: 110 ml ethanol (70%), 90 ml denaturation solution, 110 ml wash buffer, 110 ml high-purity water and 180 ml high-purity water.
- Place the vacuum prep tool (VPT) in the water trough and switch on the vacuum pump (Figure 1).
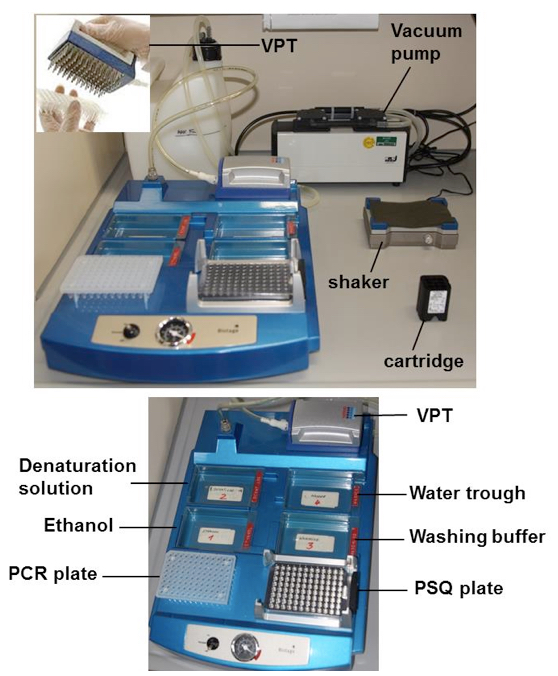
Figure 1. Vacuum workstation with the vacuum prep tool (VPT) and the different troughs
- Flush the filter probes for 20 sec.
- Move the VPT into the PCR plate to capture the beads containing immobilized templates
- Transfer the VPT sequentially to the troughs containing:
- 70% ethanol for 5 sec.
- Denaturation solution for 5 sec.
- Washing buffer for 5 sec.
- 70% ethanol for 5 sec.
- Raise the VPT at a 90° angle for 5 sec to drain liquid from the filter probes.
- Switch off the vacuum pump and return the VPT the horizontal position.
- Place the VPT in the PSQ 96 plate to release the beads. Shake the VPT gently and let it rest for a few seconds on the bottom of the wells.
- Move the VPT to the water trough to wash the filters by agitating for 10 sec.
- Heat the PSQ plate at 80 °C for 2 min to denature the templates. Cool the PSQ plate at room temperature for at least 5 min to allow the annealing of the sequencing primer.
- Select the instrument parameters according to the manufacturer's instructions, including the “sequence to analyze” obtained in the step 1g. Load the cartridge with the estimated amount of PSQ reagents.
- Place the plate in the PSQ 96 instrument and insert the cartridge carefully. Start the run
- At the end of the run, a pyrogram is shown in a graphical display (Figure 2) and the PSQ software assigns a genotype to each sample by comparison to a previously defined theoretical pattern based on the “sequence to analyze”.
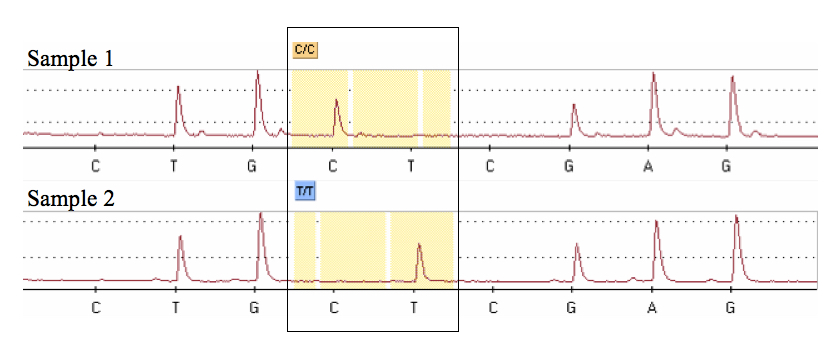
Figure 2. Pyrograms obtained after the pyrosequencing analysis showing each of two possible genotypes at a T/C SNP
Notes
- Only tailed forward primers were tested in this protocol. The tailing of reverse primers should work in the same efficient way.
- If a clear and strong PCR product is obtained after amplification, the 25 µl of reaction mix might be reduced to only 15 µl, without any negative effect on the subsequent pyrosequencing analysis.
- Various primer stoichiometries (1:2, 1:5, 1:10; tailed-forward primer: universal primer) were checked to assess which concentration of universal primer produced the highest amount of biotynilated PCR fragments. No differences were observed in the pyrograms based on the different ratios of tailed forward and universal primers. The 1:10 proportion was selected as it allows to decrease the expense of labelled primer (Silvar et al., 2011).
Recipes
- Annealing buffer
20 mM Tris-acetate
2 mM Mg-acetate
pH 7.6
- Binding buffer
10 mM Tris-HCl
2 M NaCl
1 mM EDTA
0.1% Tween 20
pH 7.6
- Denaturation solution
0.2 M NaOH
- Washing buffer
10 mM Tris-acetate
pH 7.6
- Gel running buffer
1x Tris-Borate-EDTA
89 mM Tris base
89 mM boric acid
2 mM EDTA (pH 8.0)
- 6x DNA loading buffer
30% (v/v) glycerol
0.25% (w/v) bromophenol blue
0.25% (w/v) xylene cyanol FF
Acknowledgments
This protocol is adapted from Silvar et al. (2011). The pyrosequencing assay, including annealing plate preparation, immobilization of PCR products to streptavidin beads and the preparation of single-stranded pyrosequencing template DNA were basically carried out as described in the manufacturer´s instructions.
References
- Schuelke, M. (2000). An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol 18(2): 233-234.
- Silvar, C., Perovic, D., Casas, A. M., Igartua, E. and Ordon, F. (2011). Development of a cost effective pyrosequencing approach for SNP genotyping in barley. Plant Breed 130(3): 394-397.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Silvar, C., Perovic, D., Casas, A. M., Igartua, E. and Ordon, F. (2015). Pyrosequencing Approach for SNP Genotyping in Plants Using a M13 Biotinylated Primer. Bio-protocol 5(10): e1473. DOI: 10.21769/BioProtoc.1473.
Category
Plant Science > Plant molecular biology > DNA > Genotyping
Molecular Biology > DNA > Genotyping
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









