- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantification of HIV RNA and Human Herpesvirus DNA in Seminal Plasma
Published: Vol 5, Iss 9, May 5, 2015 DOI: 10.21769/BioProtoc.1465 Views: 11124
Reviewed by: David PaulValeria LullaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A SYBR Green-based Real Time RT-PCR Assay for Detection of the Emerging H7N9 Virus
Zheng Zhu and Lunbiao Cui
Jun 20, 2014 10098 Views

Infectious Virus Yield Assay for Hepatitis E Virus
Yannick Debing [...] Johan Neyts
Aug 5, 2014 11471 Views

Quantification of HIV-2 DNA in Whole Blood
Zsófia Szojka [...] Patrik Medstrand
Oct 20, 2019 6004 Views
Abstract
Multiple viruses can co-infect the genital tract, modifying the immunologic and virologic milieu and possibly playing a role in viral transmission and pathogenesis. The aim of our studies has been to understand the complex relationships between HIV-1 RNA, and multiple human herpesviruses known to frequently replicate in the genital tract of HIV-infected men (i.e. cytomegalovirus [CMV], Epstein Bar virus [EBV], herpes simplex virus [HSV] types 1 and 2, and human herpesviruses [HHV] 6, 7 and 8) (Gianella et al., 2013a; Gianella et al., 2013b; Gianella et al., 2013c; Gianella et al., 2014). This protocol was designed to collect and process male genital secretion (GS), and to isolate and further quantify HIV RNA and DNA of seven HHV from seminal plasma using quantitative real time PCR technology.
Keywords: Seminal PlasmaMaterials and Reagents
- For male genital secretion (GS) processing
- Gibco® Hank’s balanced salt solution (HBSS) (no calcium, no magnesium, no phenol red) (Life Technologies, catalog number: 14175-095 )
- Gibco® RPMI 1640 medium (Life Technologies, catalog number: 11875-093 )
- GemCellTM U.S. origin fetal bovine serum (FBS) (Gemini-Bio Products, catalog number: 100-500 )
- Gibco® penicillin-streptomycin (10,000 U/ml) (Life Technologies, catalog number: 15140-122 )
- Gibco® L-Glutamine (200 mM) (Life Technologies, catalog number: 25030-024 )
- Nystatin suspension (10,000 units/ml in DPBS) (Sigma-Aldrich, catalog number: N1638 )
- Dimethyl sulfoxide (Hibri-MaxTM, sterile-filtered) (Sigma-Aldrich, catalog number: D2650 )
- Waxie® bleach (Waxie Sanitary Supply, catalog number: 170016 )
- Viral transport medium (VTM) (see Recipes)
- 20% GS complete RPMI medium (see Recipes)
- 20% GS freeze medium (see Recipes)
- For RNA and DNA extraction and cDNA generation
- QIAamp DNA mini kit (250 or 50 QIAamp mini spin columns) (QIAGEN, catalog numbers: 51306 or 51304 )
- High pure viral RNA kit (Roche Diagnostics, catalog number: 11858882001 )
- InvitrogenTM SuperScript® III first-strand synthesis system (Life Technologies, catalog number: 18080-051 )
- Seal-Rite 1.5 ml microcentrifuge (flip-top) tube (natural) (USA Scientific, catalog number: 1615-5500 )
Note: These tubes are autoclaved before use. - Micro tubes 1.5 ml (type D, without skirted base and with assembled neutral screw cap) (SARSTEDT AG, catalog number: 72.692.005 )
- Applied Biosystems® MicroAmp® reaction tube with cap (0.2 ml) (Life Technologies, catalog number: N8010540 )
- 1x Dulbecco’s phosphate buffered saline (DPBS) (Corning, CellgroTM, catalog number: 21-031-CV )
- EDTA (0.5 M pH 8.0, molecular biology grade) (Promega Corporation, catalog number: V4231 )
- Molecular biology grade sterile purified water (RNase, DNase, proteinase free) (Corning, CellgroTM, catalog number: 46-000-CM )
- Tris hydrochloride (1 M solution pH 8.0, molecular biology grade) (Thermo Fisher Scientific, catalog number: BP1758-500 )
Note: 1 M Tris requires to be diluted to 10 mM with molecular grade water. - Ethyl alcohol pure (200 proof molecular biology grade) (Sigma-Aldrich, catalog number: E7023 )
- MgCl2 (50 mM solution) (Bio-Rad Laboratories, catalog number: 170-8872 )
- For quantitative Real Time PCR (RT-qPCR)
- DNA plasmids or amplicons to be used as standards for RT-qPCR
Note: Quantification standards for the different herpes viruses were obtained using DNA plasmid preparations with known concentrations. We provided the nucleotide sequences for each standard amplicon (in a separate fasta file from 5’ to 3’) (please see Supplementary Material). Oligonucleotides can be custom synthetized (e.g. Ultramer® Oligonucleotides, Integrated DNA technologies [IDT]) and used directly as quantification standard, after measuring the amount of PCR-component template by spectrophotometry or droplet digital PCR.
If desired, amplicons can also be inserted into a plasmid using standard kits (e.g. Zero Blunt® TOPO® PCR Cloning Kit, catalog number: K2800J10) and stocks of construct can be generated with large batch cultures (using standard protocols). After purification, quantify plasmid concentration by spectrophotometry (or ddPCR) and extract clone vectors using standard procedures. Consider linearization to increase efficiency and avoid supercoil formation. - HIV RNA standard can be obtained from the DAIDS Virology Quality Assurance (VQA) Program (https://www.aidsreagent.org/, 150,000 copies HIV-1 RNA/ml spiked into negative plasma, catalog number: 3443 )
- Molecular biology grade sterile purified water (Corning, CellgroTM, catalog number: 46-000-CM)
- Primers and probes (Table 1)
- DNA plasmids or amplicons to be used as standards for RT-qPCR
Equipment
- For male genital secretion (GS) processing
- Fisherbrand™ polyethylene hinged-lid containers (Thermo Fisher Scientific, catalog number: 03-405-40 )
- Curwood Parafilm MTM laboratory wrapping film (4-inch W X 125-ft L) (Thermo Fisher Scientific, catalog number: 13-374-10 )
- Fisherbrand™ Infecon™ specimen transport bags (Thermo Fisher Scientific, catalog number: 19-287-215 )
- Corning® 15 ml centrifuge tube (sterile) (Corning, catalog number: 430052 )
- FisherbrandTM sterile polystyrene disposable serological pipets with magnifier stripe for 1 ml, 2 ml, 5 ml, 10 ml (Thermo Fisher Scientific, catalog numbers: 13-676-10G , 13-675-3C , 13-676-10H , 13-676-10J )
- VWR® disposable aspirating pipets (polystyrene, sterile) (VWR International, catalog number: 414004-264 )
- Micro tubes (2.0 ml, Type I, with skirted base and assembled neutral screw cap) (SARSTEDT AG, catalog number: 72.694.006 )
- 2.0 ml Ext FS CryoElite sterile cryogenic vials (yellow-cap) (Wheaton Science Products, catalog number: W985866 )
- Nalgene® Mr. FrostyTM freezing container (Thermo Fisher Scientific, catalog number: 5100-0001 )
- Allied® 1,500 ml disposable collection canisters (Allied Healthcare Products, catalog number: 20-08-0003 )
- SorvallTM RC4 General Purpose Floor Model Centrifuge (Thermo Fisher Scientific, catalog number: 75004481 )
- Original Pipetman Aid® Pipet Controller (Drummond Scientific Company, catalog number: 4-000-111-TC )
- Stericup-HV, 0.45 µm, PVDF (500 ml) (EMDMILLIPORE, catalog number: SCHVU05RE )
- Stericup-GV, 0.22 µm, PVDF (500 ml) (EMDMILLIPORE, catalog number: SCGVU05RE )
- For RNA and DNA extraction and cDNA generation
- Fisher® vortex genie 2, analog vortex mixer (Thermo Fisher Scientific, catalog number: 02-215-365 )
- VWR® digital dry block heaters (or 2 heat blocks) (VWR Scientific Products, catalog number: 12621-088 )
- Allegra® 64R high performance Benchtop centrifuge (Beckman Coulter, item number: 367585 )
- Eppendorf® centrifuge (model: 5430 , 120 V, with Rotor, model: FA-45-30-11) (Eppendorf, 120V no longer available only 230V catalog number: 5427 000.216)
- Applied Biosystems® GeneAmp® PCR system 9700 (96-well gold-plated) (Life Technologies, catalog number: 4314878 )
- For quantitative Real Time PCR (RT-qPCR)
- Applied Biosystems® MicroAmp® Optical 96-Well reaction plate (Life Technologies, catalog number: N8010560 )
- Applied Biosystems® MicroAmp® optical adhesive film (Life Technologies, catalog number: 4311971 )
- Applied Biosystems® MicroAmp® 96-Well Tray/Retainer Set (Life Technologies, catalog number: 403081 )
- Corning® 50 ml centrifuge tube (sterile) (Corning, catalog number: 430290 )
- Applied Biosystems® TaqMan® environmental master mix 2.0 (200 reactions) (Life Technologies, catalog number: 4396838 )
- Applied Biosystems® 7900HT fast Real-Time PCR system with Fast 96-Well block module (Life Technologies, catalog number: 4351405 )
Procedure
- Protocol for collection and processing of male genital secretions
- After cleaning hands and penis with sanitary wipes, semen is collected by masturbation into a Fisherbrand™ Polyethylene Hinged-Lid Container with about 2 ml of VTM (after 72 h of sexual abstinence).
- Following masturbation, the semen specimen should sit for at least 30 min (but no more than 4 h) at room temperature prior to processing to allow liquefaction.
- Record volume, time of collection and start time of processing.
- Transfer ejaculate (inclusive VTM) to a sterile Corning® 15 ml centrifuge tube.
- Centrifuge at 700 x g room-temp for 12 min in the SorvallTM RC4 floor centrifuge.
- If the GS specimen is very sticky/ goopy it is recomended to break up the specimen by pipetting the sample with a sterile narrow tip opening. Pipet 5-10 times gently, holding the tip very close to the bottom of the 15 ml conical tube. Repeat centrifugation step in order to separate plasma from cells.
- All of the plasma should be collected without disrupting the cell pellet in 0.5-1.5 ml pre-labelled aliquots (SARSTEDT 2.0 ml Micro Tubes) and stored at -80 °C.
- Flick to break up the cell pellet (no vortex) and re-suspend in 10 ml of Hank’s Balanced Salt Solution.
- Centrifuge at 700 x g room-temp for 12 min to pellet cells.
- Aspirate off the HBSS wash using a sterile VWR® Disposable Aspirating Pipet and Allied® 1,500 ml Disposable Collection Canister (vacuum trap) with 10% Waxie® Bleach.
- Flick to break up the cell pellet (no vortex) and re-suspend seminal cells in 1 ml of 20% complete RPMI medium for GS.
- Pipet a few times to gently homogenize the seminal cells in 1 ml of 20% GS complete RPMI medium without vortexing.
- Add 1 ml of 20% GS freeze medium to bring the total volume to ~2 ml and pipet a few times to mix evenly again no vortexing. Keep the cells on ice at all times.
- Aliquot the 2 ml cell suspension into 4 x 500 µl aliquots (yellow-cap, 2.0 ml CryoElite Sterile Cryogenic Vials) and freeze viably in Nalgene® Mr. FrostyTM Freezing Container* at -80 °C overnight. The next day, move to -150 °C liquid nitrogen storage.
Please note other cell cryopreservation methods/canisters are acceptable as long as the cells are viably frozen slowly at -1 °C per min. Collection and storage of seminal cells (steps A7-13) is optional. Stored cells can be used for future studies but will not be used for this protocol.
- Isolation of nucleic acids
- Thaw 1 ml of seminal plasma at room temperature.
- Immediately transfer 0.2 ml of seminal plasma into 1.5 ml SARSTEDT screw cap tube containing 20 μl of 0.5M EDTA solution to inhibit DNAase activity. Mix well by pipetting, quickly vortex, and put on ice. Repeat this step to obtain a duplicate of the same sample. Then extract DNA as described below.
Note: Duplicates are only necessary to run the entire panel of seven HHV. - In preparation for RNA extraction quickly transfer 0.5 ml of seminal plasma into a 1.5 ml SARSTEDT screw cap tube containing 0.5 ml 1x DPBS and pipet gently a few times to reduce viscosity. Then immediately put on ice.
- To concentrate RNA, centrifuge the 1 ml seminal samples with DBPS at 4 °C for 1 h at 23,500 x g using the Allegra® 64R centrifuge. Refer to RNA extraction described below.
Note: Do not allow the samples to sit around after the high-speed spin; the viral pellet is unstable!
For RNA extraction (high pure viral RNA kit)
- Following centrifugation, carefully remove the supernatant from each tube leaving 200 μl, not disturbing the invisible RNA pellet.
- Extract RNA following the manufacture’s protocol (high pure viral RNA kit).
Note: To increase RNA yield, incubate for 10 min at room temperature after adding the binding buffer to the sample. To further increase yield, incubate High Pure Filter Tube loaded with 50 μl heated elution buffer for 5 min. Heat elution buffer at 60 °C before elution step. - After elution, transfer RNA extracts to pre-labelled 1.5 ml SARSTEDT screw cap tubes and put on ice. Immediately proceed to cDNA generation discussed below. Finally store remainder of RNA extract at -80 °C.
For DNA extraction (QIAamp DNA mini kit)
- Extract DNA following the manufacturer’s protocol (QIAamp DNA mini kit).
Note: To increase DNA yield incubate the QIAamp Mini column loaded with 100 μl heated AE Buffer for 10 min. Heat AE buffer at 60 °C before elution step. - Centrifuge for 1 min at max speed (16,000 x g) in a clean seal-Rite 1.5 ml microcentrifuge tube (flip-top).
- Combine identical duplicates respectively and transfer to pre-labelled 1.5 ml SARSTEDT screw cap tube to be stored at 4 °C short-term or -20 °C long-term storage.
For cDNA HIV-1 generation (InvitrogenTM SuperScript® III first-strand synthesis system)- In a 0.2 ml MicroAmp® reaction tube with cap, mix 10 μl of extracted RNA (for seminal sample and HIV RNA for quantification standard) with 1 μl of 20 μM HIV-1 reverse primer (see Table 1) and 1 μl of 10mM dNTPs (primers and dNTPs can be pre-combined).
- Incubate at 65 °C for 5 min.
- Place immediately on ice for at least 1 min.
- To each reaction tube add (can be pre-mixed):
- 2 μl of 10x RT buffer from kit
- 2 μl MgCl2 (50 mM) *not included in kit
- 2 μl 0.1 M DTT from kit
- 1 μl RNase OUT enzyme (40 U/ml) from kit
- 1 μl SuperScript III RT enzyme (200 U/ml) from kit
- Incubate at 50 °C for 50 min and inactivate reaction at 85 °C for 5 min.
- Store at 4 °C short-term and -20 °C long-term storage.
- Quantification of cDNA and DNA viruses by RT-qPCR
For cDNA quantification RT-qPCR- Standard preparation: Each HIV RNA standard aliquot (DAIDS VQA) contains 150,000 copies HIV-1 RNA/ml spiked into negative plasma. It is important to treat the quantification standard exactly the same way as the seminal plasma samples (inclusive RNA extraction, cDNA generation and quantification). The use of RNA standards takes the variable efficiencies of the different steps into account (especially the reverse transcription step). cDNA generated during reverse transcription will be used as the template for amplification in the subsequent RT-qPCR. Example of serial dilution from cDNA using 10 mM Tris buffer:(*5 μl in 15 μl 10 mM Tris buffer)
Standard Dilution factor Copies per well D0 Undiluted 3,750 D1 4* 938 D2 4* 234 D3 4* 59 D4 4* 15 D5 4* 4 Neg ctrl H2O 0
All standards (including negative control) should be run at least in duplicate (or triplicate if possible).
Note: Do not store quantification standards for prolonged periods of time and thaw only once before use. - In a 96 well plate, mix 5 μl of cDNA, 25 μl TaqMan® Environmental Master Mix 2.0, HIV primers (1 μM) and probe (0.3 μM) as displayed in Table 1. Add water to adjust the volume to 50 μl total volume per reaction per well. Each sample should be run in duplicate (or in triplicates) and final result will be obtained as an average value of the replicate wells.
- PCR cycling conditions:
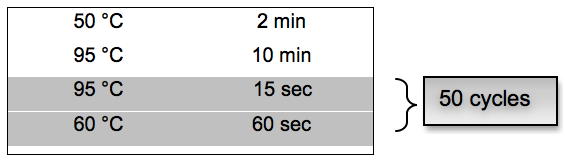
For DNA Quantification RT-qPCR- Standard preparation: Quantification standards for the different herpes viruses were obtained using plasmid preparations with known concentrations. Because plasmid sequences are highly abundant (for example 1 x 1010 - 1 x 1012 per μl), they can be sources of PCR contamination. Extreme caution must be exercised when working with these DNAs to prevent their exposure to stock PCR reagents, solvents used for the dilution of PCR reagents and laboratory equipment and surfaces. Prepare little aliquots (for example 2 μl) to avoid multiple thawing.
Example of serial dilution (using 10 mM Tris) starting from a plasmid concentration (stock=D0) of 2 x 1010 per μl. Accurate pipetting is essential because the standards must be diluted over several orders of magnitude.(*2 μl in 50 μl 10 mM Tris buffer, **20 μl in 180 μl 10 mM Tris buffer)Dilution Dilution factor Copies per µl D0 Undiluted stock 2 x 1010 D1 25* 8 x 108 D2 10** 8 x 107 D3 10** 8 x 106 D4 10** 8 x 105 D5 10** 8 x 104 - Dilution D5 can be used as the highest concentration for the actual standard curve. Starting from D5, perform a 1:10 dilution as follows (note: numbers of copies are now indicated per PCR well):(*5 μl in 45 μl 10 mM Tris buffer)
Dilution Dilution factor Copies per well
(per 10 µl)D5 10* 8 x 104 D6 10* 8 x 103 D7 10* 8 x 102 D8 10* 8 x 101 D9 10* 8 D10 10* 0.8 D11 10* 0.08 Neg ctrl H2O 0 - In a 96 well plate, mix 10 μl of DNA, 25 μl TaqMan® Environmental Master Mix 2.0, specific primers (1 μM) and probe (0.3 μM) as displayed in Table 1. Add water to adjust the volume to 50 μl total volume per reaction per well. Each sample should be run in duplicate (or in triplicates whenever possible) and final result will be obtained as an average value of the replicate wells.
- PCR cycling conditions:

- Standard preparation: Each HIV RNA standard aliquot (DAIDS VQA) contains 150,000 copies HIV-1 RNA/ml spiked into negative plasma. It is important to treat the quantification standard exactly the same way as the seminal plasma samples (inclusive RNA extraction, cDNA generation and quantification). The use of RNA standards takes the variable efficiencies of the different steps into account (especially the reverse transcription step). cDNA generated during reverse transcription will be used as the template for amplification in the subsequent RT-qPCR. Example of serial dilution from cDNA using 10 mM Tris buffer:
Analysis
After the run is completed, the CT fluorescence level line should be adjusted to fall within the steep part of the amplification curve (see Figure 1). Amount of unknown target (samples) can be extrapolated from the standard curve using the formula for linear regression: y = ax + b
a = slope obtained from the standard curve.
b = intercept obtained from the standard curve.
y = Threshold Ct value for seminal sample.
x = Extrapolated amount of target DNA or cDNA for seminal sample.
Note: Consider repeating samples with differences of 0.3 Cts between duplicates. Run triplicates whenever possible to improve precision. Samples at the lower limit of detection might have discordant Ct values due to Poisson distribution of templates across wells. Thus, for more reliable low copy detection, a large number of replicates are necessary to provide statistical significance and to overcome the Poisson distribution limitation.
Calculator template for the analysis and an example are provided in a separate excels spreadsheet (“example_calculator”). 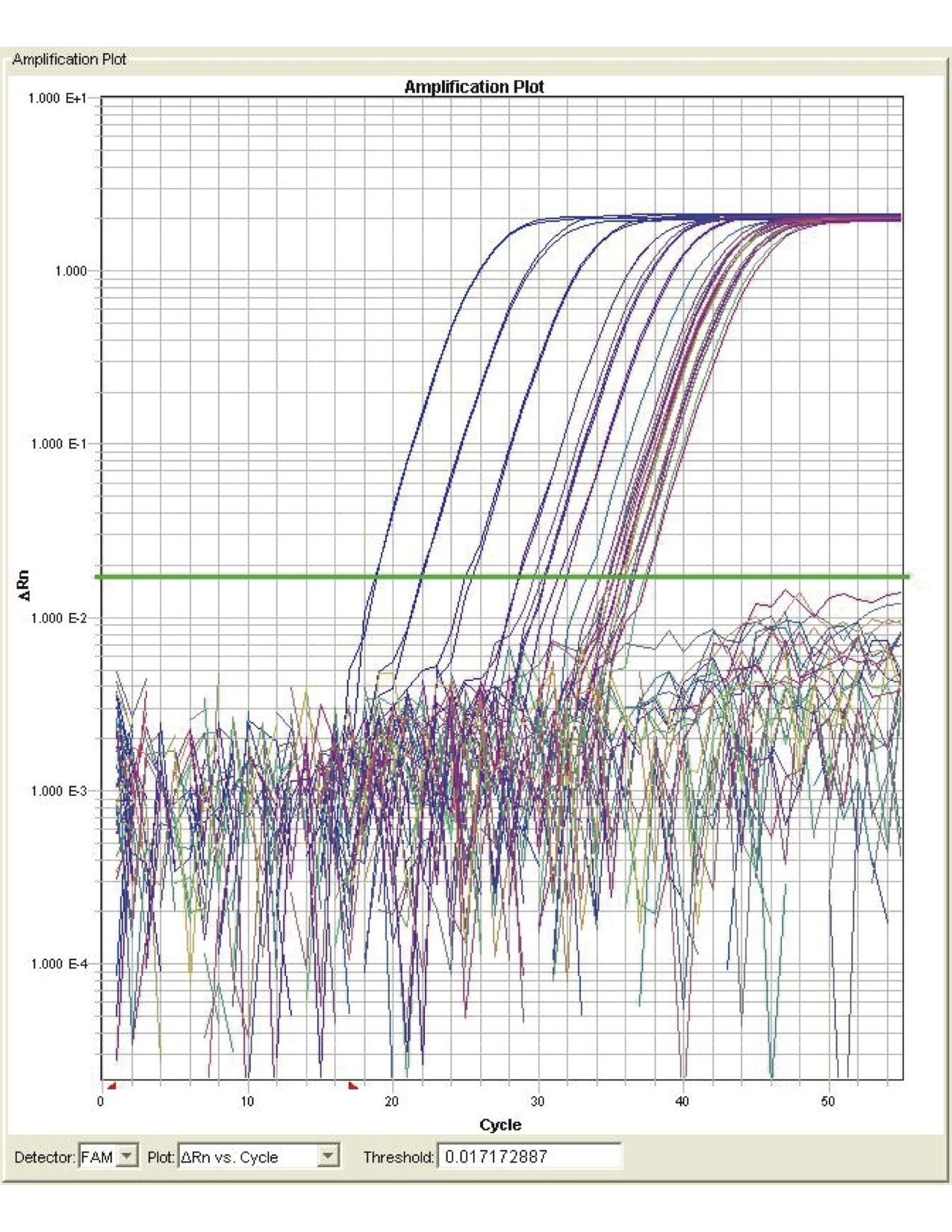
Figure 1. Amplification plot
Notes
- Donor should try not to ejaculate for at least 72 h (3 days) before producing sample.
- Donor should produce semen on the same day that he is bringing it in. Do not ejaculate the night before and store the semen in the refrigerator.
- Donor should avoid any form of lubricant or saliva during masturbation.
- Longer arousal and masturbation before ejaculation is encouraged to produce larger volume of semen (at least 15-20 min).
- When handling potentially infectious materials it’s highly recommended to use personal protective equipment/gear and universal precautions.
Recipes
Note: Recipes for processing GS Media (stored at 4 °C). GemCellTM U.S. origin fetal bovine serum (FBS) is filtered twice; once with 0.45 µm Stericup-HV and then with 0.22 µm Stericup-GV prior to GS media preparation.
- Viral transport medium (VTM)
RPMI 1640 medium
100 U/ml pen/strep
200 U/ml nystatin
2 mM L-glutamine
10% FBS filtered - 20% GS complete RPMI medium
RPMI 1640 medium
100 U/ml pen/strep
2 mM L-glutamine
20% FBS filtered - 20% GS freeze medium
20% DMSO
80% FBS filtered
Supplementary Materials
Table 1. Primers and Probes for HIV and different Herpesviruses, adapted from Gianella et al. (2013b)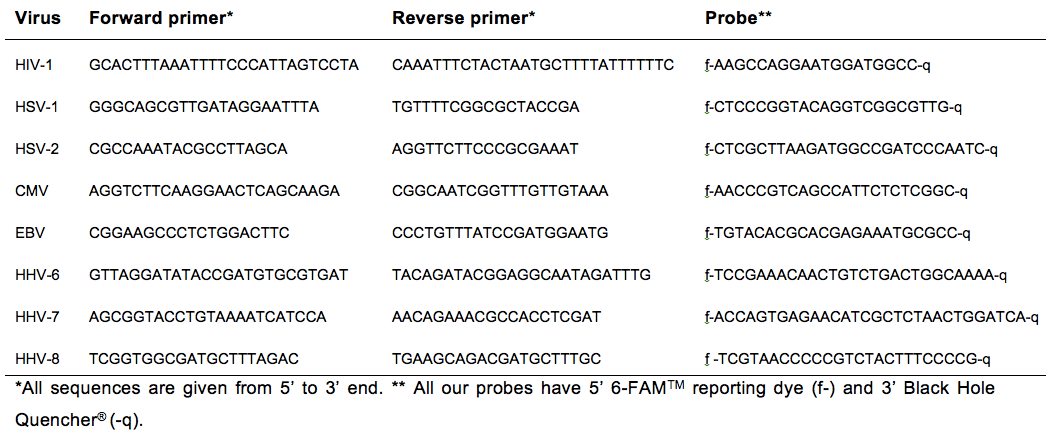
Table 2. Nucleotide sequences for each herpes viruses to use for standard plasmids or Amplicons
Acknowledgments
This work is supported by the Department of Veterans Affairs, the UCSD Center for AIDS Research (P30 AI36214), the James B. Pendleton Charitable Trust, AmfAR grant 108537 with support from FAIR, a CTRI pilot grant: UL1TR000100, and National Institutes of Health (NIH) awards: P30-AI027763, MH101012, 7-UM1 AI068636-07. A special appreciation goes to our belated Marek Fisher, Matt Strain, Antoine Chaillon and Stephen Espitia for all his help and input in developing and validating this protocol.
References
- Gianella, S., Morris, S. R., Anderson, C., Spina, C. A., Vargas, M. V., Young, J. A., Richman, D. D., Little, S. J. and Smith, D. M. (2013a). Herpesviruses and HIV-1 drug resistance mutations influence the virologic and immunologic milieu of the male genital tract. AIDS (London, England) 27(1): 39.
- Gianella, S., Smith, D. M., Vargas, M. V., Little, S. J., Richman, D. D., Daar, E. S., Dube, M. P., Zhang, F., Ginocchio, C. C., Haubrich, R. H., Morris, S. R. and Team, C. (2013b). Shedding of HIV and human herpesviruses in the semen of effectively treated HIV-1-infected men who have sex with men. Clin Infect Dis 57(3): 441-447.
- Gianella, S., Morris, S. R., Vargas, M. V., Young, J. A., Callahan, B., Richman, D. D., Little, S. J. and Smith, D. M. (2013c). Role of seminal shedding of herpesviruses in HIV Type 1 Transmission. J Infect Dis 207(2): 257-261.
- Gianella, S., Massanella, M., Richman, D. D., Little, S. J., Spina, C. A., Vargas, M. V., Lada, S. M., Daar, E. S., Dube, M. P., Haubrich, R. H., Morris, S. R., Smith, D. M. and California Collaborative Treatment Group, T. (2014). Cytomegalovirus replication in semen is associated with higher levels of proviral HIV DNA and CD4+ T cell activation during antiretroviral treatment. J Virol 88(14): 7818-7827.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Vargas-Meneses, M. V., Massanella, M., Ignacio, C. C. and Gianella, S. (2015). Quantification of HIV RNA and Human Herpesvirus DNA in Seminal Plasma. Bio-protocol 5(9): e1465. DOI: 10.21769/BioProtoc.1465.
Category
Microbiology > Microbial genetics > RNA > qRT-PCR
Microbiology > Microbial genetics > DNA > DNA quantification
Microbiology > Microbe-host interactions > Virus
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











