- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantification of Ralstonia solanacearum Colonization of Potato Germplasm Using Luminescence
Published: Vol 5, Iss 9, May 5, 2015 DOI: 10.21769/BioProtoc.1461 Views: 11172
Reviewed by: Arsalan DaudiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Quantification of the Composition Dynamics of a Maize Root-associated Simplified Bacterial Community and Evaluation of Its Biological Control Effect
Ben Niu and Roberto Kolter
Jun 20, 2018 10488 Views

Tracking Root Interactions System (TRIS) Experiment and Quality Control
Hassan Massalha [...] Asaph Aharoni
Apr 20, 2019 7336 Views

A Quick Method for Screening Biocontrol Efficacy of Bacterial Isolates against Bacterial Wilt Pathogen Ralstonia solanacearum in Tomato
Heena Agarwal [...] Niraj Agarwala
Nov 20, 2020 5547 Views
Abstract
We have developed an unsophisticated, non-disruptive and accurate method for evaluation of pathogen colonization in planta. In this protocol we use a Ralstonia solanacearum (R. solanacearum) UY031 strain genetically modified to constitutively generate light from a synthetic luxCDABE operon stably inserted in its chromosome. This system allows bacterial quantification in a high-throughput manner, avoiding time-consuming and tedious bacterial dilution plating and colony counting. In addition, this system could be especially useful in plant breeding programs to detect bacterial latent growth in symptomless parental lines before their inclusion in long-term disease resistance breeding programs.
Materials and Reagents
- Ralstonia solanacearum UY031 strain transformed with the reporter plasmid pRCG-Ppslux, which bears the chloroplast promoter PpsbA and the entire LuxCDABE operon (Figures 1 and 2)
Note: Plasmid and strain available under material transfer agreement (MTA) from Marc Valls’ laboratory. - Four-week-old Solanum tuberosum (S. tuberosum) and Solanum commersonii (S. commersonii) plants
Note: They were grown in a TREF universal potting soil mix, in a greenhouse, with 12 h light and temperatures maintained between 22 to 25 °C (50 to 60% relative humidity -RH) for three weeks and then transferred for an additional week into a growth chamber at 27 °C and 65% RH with a photoperiod of 12 h of light. - Gentamicin (75 µg/ml in solid and 5 µg/ml in liquid cultures) (Sigma-Aldrich, catalog number: G1264 )
- Bacto-peptone (BactoTM, catalog number: 211677 )
- Yeast extract (BactoTM, catalog number: 210929 )
- Tryptone (BactoTM, catalog number: 211699 )
- Casamino acids (BactoTM, catalog number: 223050 )
- Dextrose Glucose (DifcoTM, catalog number: 215510 )
- Bacto-agar (BactoTM, catalog number: 214030 )
- Triphenyltetrazolium chloride (TTC) (DifcoTM, catalog number: 231121 )
- Sodium phosphate dibasic heptahydrated (Na2HPO4.7H2O) (Sigma-Aldrich, catalog number: S9390 )
- Potassium phosphate (KH2PO4) (Sigma-Aldrich, catalog number: P5655 )
- Sodium chloride (NaCl) (VWR International, catalog number: 443824T )
- Ammonium chloride (NH4Cl) (VWR International, catalog number: 0621 )
- Magnesium sulfate (MgSO4) (VWR International, catalog number: 0338 )
- Calcium chloride (CaCl2) (VWR International, catalog number: 1.02391.1000 )
- L-glutamate (Sigma-Aldrich, catalog number: G1626-100G )
- Rich B medium (see Recipes)
- Boucher’s minimal medium (MM) (see Recipes)
- Luria and Bertani broth (LB) (see Recipes)
- 1 M magnesium sulfate (MgSO4) (see Recipes)
- 20% glucose (see Recipes)
- 1 M calcium chloride (see Recipes)
- 20 mM L-glutamate (see Recipes)
- To prepare 1 L of 1x MM (see Recipes)
Equipment
- LAS4000 Chemiluminescence and Fluorescence Imaging System (Fujifilm Life Science)
- Luminometer Berthold FB 12 (Titertek-Berthold, catalog number: 11010102 )
- Spectrophotometer (Shimadzu Kyoto, model: UV-1603 visible spectrophotometer)
- Incubator that can be set at 30 °C containing shaker for Erlenmeyer and tubes
- Tabletop centrifuge (Eppendorf, model: 5418R )
- Culture tubes (overflow volume 22 ml) (VWR International, catalog number: 47729-580 )
- Sterile 150 ml flasks
- 2 ml Eppendorf tubes
- Razor blade
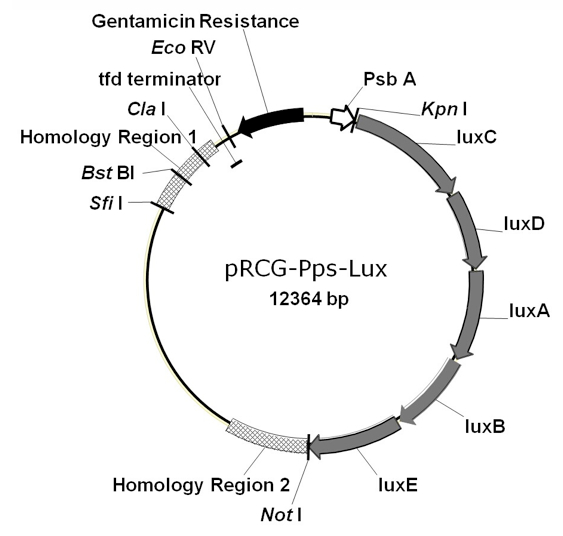
Figure 1. Vector map of pRCG-Pps-lux, designed to generate the luminescent strain for detection of bacterial colonization and growth in planta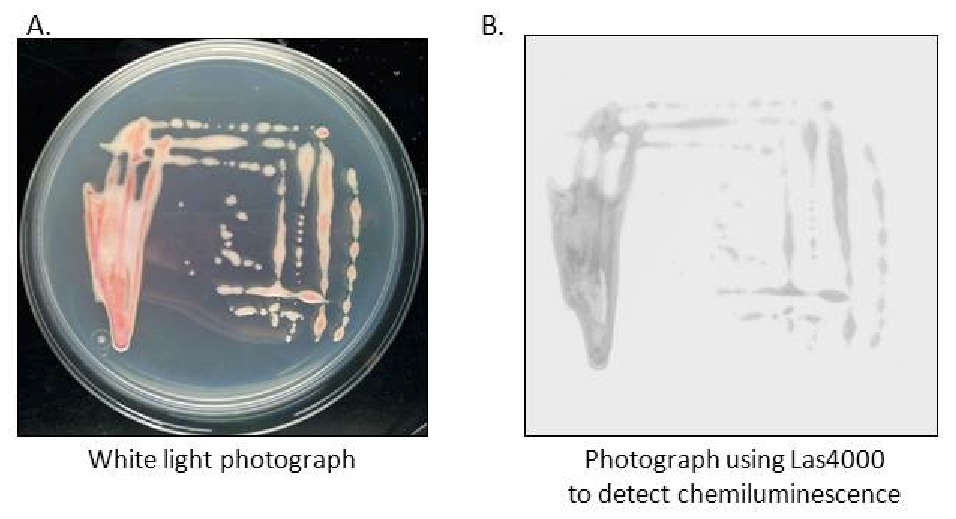
Figure 2. Visualization of Ralstonia solanacearum transformed with the pRCG-Pps-lux plasmid under white light A or chemiluminiscence B using the LAS4000 light imager
Procedure
- Measuring lux reporter expression in culture
- Plate bacterial strain from -80 °C frozen stock in a petri plate containing solid rich B medium, with TTC and selecting antibiotics (in this case gentamicin) overnight at 30 °C.
- Next day, get one colony from the plate, and grow bacterial strain overnight in a culture tube with 5 ml of rich B liquid medium (do not add TTC) in a shaker at 30 °C.
- The next day, make a ¼ dilution of the bacterial culture and measure its concentration using a UV visible spectrophotometer.
- Prepare minimal medium (MM) (20 ml per sample) into 150 ml flasks.
- Dilute overnight bacterial cultures in fresh MM adjusting bacterial concentration to a final OD600 of 0.1 in MM (approximately 108 cells/ml).
- Grow bacterial cultures at 30 °C with shaking at 200 rpm.
- Take 1 ml samples of the bacterial cultures every hour during 8 h starting at 0 h (control) for a total of 9 samples.
- For each sample, measure bacterial OD600 using the UV visible spectrophotometer and luminescence using a luminometer (express as Relative Luminescence Units or RLU). We have not used different luminometers; however, using different machines should not have an effect, because what matters are the relative measurements. Each machine should be calibrated with the control (not inoculated tissue) and all subsequent measurements are made with respect to the control for that machine.
- Establish background levels of luminescence.
- Create a correlation between bacterial concentration OD600 and RLU units (Figure 3).
Figure 3. Correlation between cell number and luminescence in in vitro cultures of UY031 Pps-lux. Overnight cultures of the bacterial strain carrying the PpsbA::luxCDABE gene fusion were diluted in Minimal Medium and bacterial growth and reporter output were measured, respectively through OD600 and luminescence emission of 1 ml aliquots taken over an eight hour time-course. RLU= Relative luminescence units
- Plate bacterial strain from -80 °C frozen stock in a petri plate containing solid rich B medium, with TTC and selecting antibiotics (in this case gentamicin) overnight at 30 °C.
- Plant inoculation and disease rating
- Plate bacterial strain carrying the pRCG-Pps-lux plasmid, from -80 °C frozen stock in a petri plate containing solid rich B medium, with TTC and selecting antibiotics (in this case gentamicin) overnight at 30 °C.
- Next day, get one colony from the plate and grow bacterial strain overnight in a 20 ml culture tube with 5 ml of rich B liquid medium (do not add TTC) in a shaker at 200 rpm at 30 °C.
- Pellet bacterial cells by centrifugation on a tabletop centrifuge at 5,914 x g for 2 min.
- Re-suspend the bacterial cells in 2 ml of water and spectrophotometrically adjust the concentration to 100 CFU/ml (corresponding to 0.01 OD600).
- Slightly damage the roots of four-week-old plants (S. commesonii and S. tuberosum in our case) by making three holes in the soil of each pot with a 1-ml pipette tip (2 cm deep) around the plant stem prior to inoculation. See Video 1 for more detail.Video 1. Drench inoculation of potato with Ralstonia solanacearum
- Drench-inoculate plants with 40 ml of a bacterial suspension at a concentration of 100 CFU/ml (corresponding to 0.01 OD600) per 12 cm 3 pots.
- After inoculation, keep plants in a growth chamber at 27 to 28 °C (65% relative humidity) with a 12 h photoperiod.
- Record disease development daily using an ordinal disease index scale ranging from 0 (no wilting symptoms) to 4 (all leaves wilted) (Winstead and Kelman, 1952) (Figure 4).

Figure 4. Visual assessment of disease development by rating plant wilting phenotype: zero: No wilting; two: 50% of plant wilted; four: All leaves wilted (Zuluaga et al., 2014)
- Plate bacterial strain carrying the pRCG-Pps-lux plasmid, from -80 °C frozen stock in a petri plate containing solid rich B medium, with TTC and selecting antibiotics (in this case gentamicin) overnight at 30 °C.
- Bacterial visualization in planta
Bacterial visualization: Non-disruptive method using LAS4000 light imager system.- Inoculate plants as described above.
- Visualize bacterial colonization of the whole plants using a Fujifilm LAS4000 light imager system with the following settings: Chemiluminescence method, incremental exposure time of 2 min, sensitivity/resolution set to high binning (Figure 5).
- Evaluate plants daily to assess for pathogen colonization.
Figure 5. Bacterial colonization of whole plants using the non-disruptive method with the LAS4000 light imager system (Zuluaga et al., 2014). Days after inoculation= dai - Example of the applicability of the protocol: Using bacterial visualization in planta as a tool to select for resistant potato genotypes against R. solanacearum.
Visualization of pathogen colonization in planta using LAS4000: Correlation between disease symptoms and bacterial colonization of the plant tissue is not always direct, due to potential latent growth. The pictures below illustrate that some plants with symptoms ratings of zero (no disease) are colonized by bacteria (Figure 6). Using a bacterial strain transformed with the pRCG-Pps-lux plasmid could be especially useful in plant breeding to detect latency in symptomless parental lines before their inclusion in long-term disease resistance breeding programs.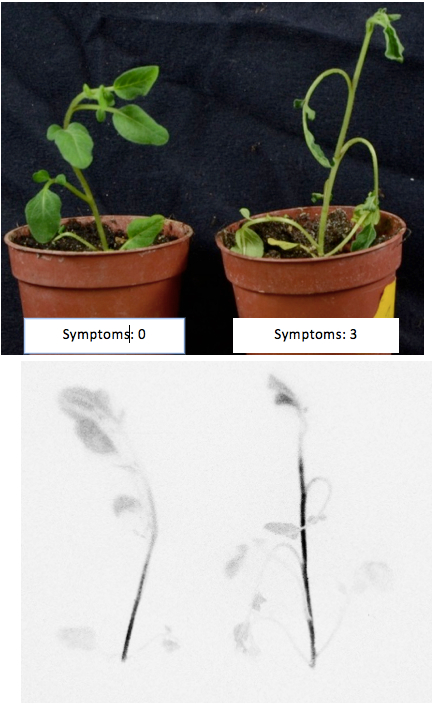
Figure 6. High-throughput non-disruptive evaluation of potato germplasm using R. solanacearum Lux::CDABE strain to assist selection of resistant genotypes
- Inoculate plants as described above.
- Bacterial quantification: Disruptive method using a luminometer (Berthold FB 12).
- To quantify bacteria present in plants in a high-throughput manner, and avoid bacterial dilution plating and colony counting, collect one-cm sections of the root system or the stem from 1 cm above or below ground depending on the tissue to evaluate (1 cm below ground in this case, since we are evaluating root colonization).
- Introduce cut section (1 cm root tissue) in a 2 ml Eppendorf tube.
- Measure luminescence directly from the cut sections with a luminometer (Berthold FB 12).
- Weight each sample.
- Correct luminescence readings for the amount of tissue present in each sample to express the final values in RLU per milligram of tissue (Figure 7).
- Use the RLU/OD600 curve generated (Figure 3) to estimate bacterial concentration.
Figure 7. Pathogen colonization is presented for four different potato genotypes with contrasting resistance to R. solanacearum. Tissue colonization is measured as Relative Luminescence Units (RLU) divided by the wet weight of the roots in milligrams (mg). Non-inoculated (N. I.) plant was used as a negative control.
- To quantify bacteria present in plants in a high-throughput manner, and avoid bacterial dilution plating and colony counting, collect one-cm sections of the root system or the stem from 1 cm above or below ground depending on the tissue to evaluate (1 cm below ground in this case, since we are evaluating root colonization).
Notes
- This procedure is highly reproducible; however, because bacterial penetration and colonization of plants is a stochastic event, at least 15 plants per treatment should be inoculated to get reliable results.
- Slight damage of the roots, by making three holes in the soil with a 1 ml pipette tip (two-cm deep) around the plant stem is highly recommended before inoculation, since bacteria need an entry point. See Video 1 for more details.
Recipes
- Rich B medium
Bacto-peptone 10 g
Yeast extract 1 g
Casamino acids 1 g
Glucose 5 g
Solid medium contained Bacto-agar 15 g and TTC 50 mg
Add distilled water to 1 L and autoclave - Boucher’s minimal medium (MM) (Boucher et al., 1985; Monteiro et al., 2012)
5x M9 saline solution stock
Dissolve in 800 ml of distilled water
Sodium phosphate heptahydrated (Na2HPO4.7H2O) 64 g
Potassium phosphate (KH2PO4) 15 g
Sodium chloride (NaCl) 2.5 g
Ammonium chloride (NH4Cl) 5.0 g
Stir until dissolved and then adjust volume to 1 L with distilled water and autoclave - Luria and Bertani broth (LB) (Sambrook et al., 2000)
Yeast extract 5 g
Tryptone 10 g
NaCl 10 g
Add distilled water to 1 L and autoclave - 1 M magnesium sulfate (MgSO4)
Autoclaved - 20% glucose
Filter sterilized - 1 M calcium chloride (CaCl2)
Autoclaved - To prepare 1 L of 1x MM (the final medium where R. solanacearum is grown)
200 ml saline solution stock M9 (5x)
800 ml distilled water
Autoclave
Once the medium is autoclaved, let it cool down to 60-65 °C and then add:
2 ml of 1 M MgSO4
0.1 ml of 1 M CaCl2
20 mM L-glutamate as a carbon source (filter sterilized)
Acknowledgments
This work was supported by grants SGR0052 and CONES2010-0030 from Comissionat per Universitats i Recerca of the Catalan Government (Generalitat de Catalunya), AGL2010-21870 from the Ministerio de Economía of the Spanish Government, and FMV_2009_1_3045 from the National Research Council in Uruguay (ANII). We thank I. van DijK and F. Monteiro for constructing original plasmids (pG-Pps, pRCG-GWY and pRCGent-Peplux) used as sources of inserts for cloning, F. Vilaró and M. González for providing the S. commersonii genotypes used in this study and S. Balcells for lending us a luminometer.
References
- Boucher, C. A., Barberis, P. A. and Demery, D. A. (1985). Transposon mutagenesis of Pseudomonas solanacearum: isolation of Tn5-induced avirulent mutants. Journal of Gen Microbiol 131(9): 2449-2457.
- Monteiro, F., Genin, S., van Dijk, I. and Valls, M. (2012). A luminescent reporter evidences active expression of Ralstonia solanacearum type III secretion system genes throughout plant infection. Microbiology 158(Pt 8): 2107-2116.
- Sambrook, J., Fritsch, E. F. and Maniatis, T. (2000) Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press.
- Winstead, N. N. and Kelman, A. (1952). Inoculation techniques for evaluating resistance to Pseudomonas solanacearum. Phytopathology 42: 628-634.
- Zuluaga, A. P., Ferreira, V., Pianzzola, M. J., Siri, M. I., Coll, N. S. and Valls, M. (2014). A novel, sensitive method to evaluate potato germplasm for bacterial wilt resistance using a luminescent Ralstonia solanacearum reporter strain. Mol Plant Microbe Interact 27(3): 277-285.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Zuluaga, A. P., Coll, N. S. and Valls, M. (2015). Quantification of Ralstonia solanacearum Colonization of Potato Germplasm Using Luminescence . Bio-protocol 5(9): e1461. DOI: 10.21769/BioProtoc.1461.
Category
Plant Science > Plant immunity > Disease bioassay
Microbiology > Microbe-host interactions > In vivo model > Plant
Microbiology > Microbe-host interactions > Bacterium
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










