- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro Reconstitution Assay of miRNA Biogenesis by Arabidopsis DCL1
Published: Vol 5, Iss 8, Apr 20, 2015 DOI: 10.21769/BioProtoc.1454 Views: 10870
Reviewed by: Feng LiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

An Activity-Based Proteomics with Two-Dimensional Polyacrylamide Gel Electrophoresis (2D-PAGE) for Identifying Target Proteases in Arabidopsis Apoplastic Fluid
Sayaka Matsui and Yoshikatsu Matsubayashi
Mar 5, 2025 1975 Views

Advancing 2-DE Techniques: High-Efficiency Protein Extraction From Lupine Roots
Sebastian Burchardt [...] Emilia Wilmowicz
Oct 5, 2025 1763 Views

Effective Gene Silencing in Plants by Synthetic Trans-Acting siRNAs Derived From Minimal Precursors
Adriana E. Cisneros [...] Alberto Carbonell
Oct 20, 2025 1806 Views
Abstract
microRNAs (miRNAs) are small non-coding RNAs, regulating most if not all, biological processes in eukaryotic organisms. miRNAs are initially processed from primary transcripts (pri-miRNAs) to produce miRNA precursors (pre-miRNAs), that are further processed into miRNA and its complementary strands (miRNA/*). In Arabidopsis, and possibly other plants, the processing from pri-miRNAs to pre-miRNAs and from pre-miRNAs to miRNA/* are both implemented through Dicer-like 1 (DCL1) complexes. Recently, we demonstrated isolation of DCL1 complexes of unprecedented quality from in planta. We further successfully reconstituted DCL1 cleavage assays in vitro that were able to fully recapitulate in vivo miRNA biogenesis. Here we provide a detailed protocol of DCL1 reconstitution assays. The protocol comprises three major parts (Figure 1): 1) Preparation of pri- and pre-miRNA transcripts (Procedures A-C); 2) Purification of the recombinant Arabidopsis DCL1 machinery from Nicotiana benthamiana (N. benthamiana) through immunoprecipitation (IP) (Procedures D and E); and 3) in vitro processing of radioisotope-labeled pri- or pre-miRNAs using the isolated DCL1 complexes (Procedure F). It is our desire that the protocol be a powerful tool for the RNAi community to study mechanistic issues or to develop RNA silencing technologies.
Keywords: MicroprocessorMaterials and Reagents
- Five to six-week old plants of N. benthamiana
- Agrobacterium strain: ABI (Zhang et al., 2006)
- Plasmid [pBA-2Flag-4Myc-DCL1, pBA-6Myc-HYL1, pBA-6Myc-SE. Detailed approach could refer to the Online Methods from Zhu et al. (2013). Negative control plasmid: pBA]
- G-Tube® snap cap siliconized microcentrifuge tubes (VWR International, catalog number: 22179-004 )
- Anti-FLAG® M2 magnetic beads (Sigma-Aldrich, catalog number: M8823 )
- 3x Flag peptide (NH2-MDYKDHDGDYKDHDIDYKDDDDK-COOH) (Sigma-Aldrich, catalog number: F4799 )
- DNase (Promega Corporation, catalog number: M6101 )
- 3,000 Ci/mmol 10 mCi/ml [γ-32P]-ATP (PerkinElmer, catalog number: BLU502A100UC )
- SuperaseIn RNase inhibitor (Ambion, catalog number: N8080119 )
- T7 RNA polymerase (Ambion, catalog number: 18033019 )
- Calf intestine alkaline phosphatase (NEB, catalog number: M0290S )
- Phenol: chloroform: isoamyl alcohol (25:24:1) (Life Technologies, InvitrogenTM, catalog number: 15593049 )
- NaAc (Sigma-Aldrich, catalog number: S7670 )
- 5 M ammonium acetate (Ambion, catalog: AM9071 )
- GlycoBlue (Ambion, catalog number: AM9516 )
- Decade marker (Ambion, catalog number: AM7778 )
- Anti-c-Myc Agarose Affinity Gel (Sigma-Aldrich, catalog number: A7470 )
- EDTA-free protease inhibitor cocktail (Roche Diagnostics, catalog number: 05892953001 )
- Protease inhibitor cocktail (Sigma-Aldrich, catalog number: P2714 )
- HEPES (Sigma-Aldrich, catalog number: H3375 )
- Spermidine (Sigma-Aldrich, catalog number: S2626 )
- DTT (DL-Dithiothreitol) (Sigma-Aldrich, catalog number: 43817 )
- MgCl2 (Sigma-Aldrich, catalog number: M8266 )
- NTP (Thermo scientific, catalog number: R0481 )
- Deionized formamide (Ambion, catalog number: AM9342 )
- Bromophenol blue (Sigma-Aldrich, catalog number: B0126 )
- Xylene cyanol (Sigma-Aldrich, catalog number: X4126 )
- EDTA (Sigma-Aldrich, catalog number: V900106 )
- SDS (Sigma-Aldrich, catalog number: L3771 )
- DEPC (Sigma-Aldrich, catalog number: V900882 )
- KCl (Sigma-Aldrich, catalog number: V900068 )
- Tris-HCl (Sigma-Aldrich, catalog number: V900312 )
- Triton X-100 (Sigma-Aldrich, catalog number: V900502 )
- PMSF (Sigma-Aldrich, catalog number: 78830 )
- NaCl (Sigma-Aldrich, catalog number: V900058 )
- ATP (Thermo scientific, catalog number: R1441 )
- GTP (Thermo scientific, catalog number: R1461 )
- Glycerol (Sigma-Aldrich, catalog number: V900122 )
- 0.1 M Glycine-HCl (Sigma-Aldrich, catalog number: 55097-5ML-F )
- 5x transcription buffer (see Recipes)
- RNA loading buffer (see Recipes)
- RNA elution buffer (see Recipes)
- DEPC H2O (see Recipes)
- RNA dissolve buffer (see Recipes)
- Chloroform: isoamyl alcohol (24:1) (see Recipes)
- IP buffer (see Recipes)
- Protease inhibitor cocktail stock (see Recipes)
- TBS buffer (see Recipes)
- 3x Flag elution buffer stock (4 mg/ml) (see Recipes)
- Washing buffer (see Recipes)
- Assay buffer (see Recipes)
- Fixing buffer (see Recipes)
Equipment
- Compact UV lamp (UVP, model: UVGL-25 )
- Fluor-Coated TLC Plate (10 x 10 cm) (Ambion, catalog number: AM10110 )
- Geiger counter (Medcom, model: CRM-100 )
- Vertical electrophoresis system (Whatman, model: V15.17 )
- Thermomixer C (Eppendorf, model: 5382000023 )
- Gel dryer (Bio-Rad Laboratories, model: 583 )
- Gel analyzer Quantity One (Bio-Rad Laboratories, version: 4.6.9 )
- DynaMagTM-2 (Life technologies, model: 12321D )
- Rugged Rotator (Glas-Col, model: 099A MR1512 )
- PolyATtract® System 1000 Magnetic Separation Stand (Promega Corporation, model: Z541A )
- Phospho imager (PharosFXTM plus system)
Procedure
Note: Please see Figure 1 for a schematic diagram of the six steps described below.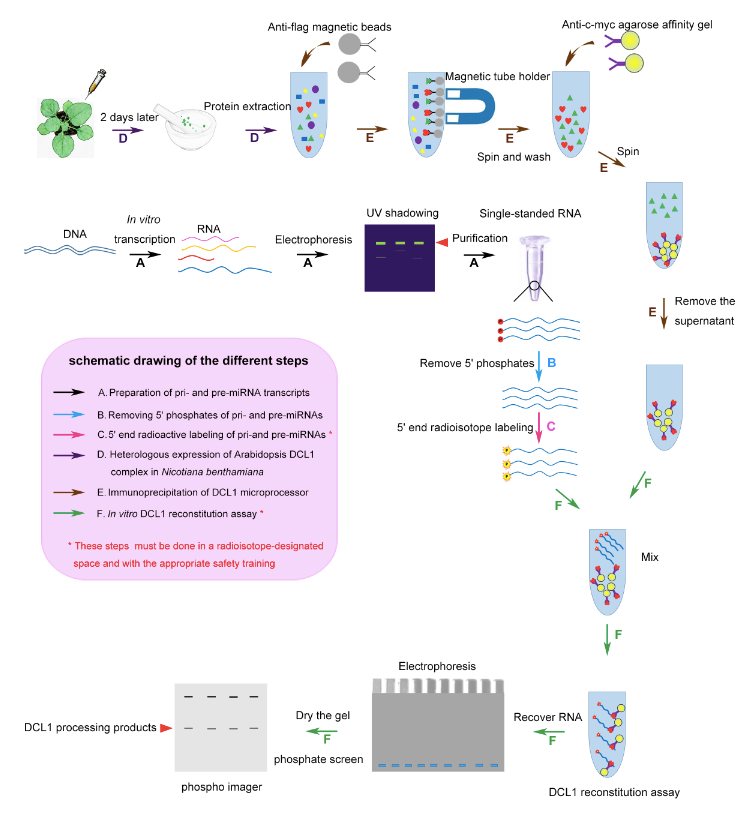
Figure 1. A schematic diagram of procedure
- Preparation of pri- and pre-miRNA transcripts (Han et al., 2014)
- Substrates of pri- and pre-miRNA are transcribed under the T7 promoter in vitro using PCR-generated templates. In this case, the forward PCR primer should contain T7 promoter sequences (TAATACGACTCACTATAG) to drive transcription directly from the PCR product.
- Prepare in vitro transcription reaction as follows:Note: In vitro transcription kits are also available from other commercial vendors.
Reagent Volume Final conc. Purified PCR product (100 ng) - µl 5x transcription buffer 4 µl Superase-In 1 µl 1 U/μl T7 RNA polymerase 2 µl Add H2O to final volume 20 µl 20 µl - Briefly spin down. Incubate at 37 °C for 3 h or overnight.
- Add 1 μl DNase. Mix and leave the tube at 37 °C for 30 min.
- Add 20 μl RNA loading buffer to the transcribed RNA.
- Heat the tubes for 2 min at 95 °C. Cool down for 2 min on ice.
- Briefly spin down the tubes at 4 °C. Keep the samples on ice until loading.
- Meanwhile, prepare a 6% urea-polyacrylamide gel.
- Before loading the samples, flush the wells of the 6% urea-polyacrylamide gel with 1x TBE running buffer to remove the diffused urea from the wells.
- Load the RNA samples on 6% urea-polyacrylamide gel and run at 350 V (20 V/cm) until bromophenol blue reaches the bottom of the gel.
- Remove one of the glass plates from the gel.
- Apply UV shadowing to locate the positions of transcripts and then cut out the gel slice of single bands of pri- or pre-miRNAs band.
UV Shadowing: Remove the gel from the glass plates used for the electrophoresis and place it on a sheet of plastic wrap. Then position the plastic wrap and gel on top of a fluor-coated TLC plate (attention: Avoid skin and eye exposure to UV light). Using a hand-held UV monitor, identify the bands that exhibit distinguishable shadow and mark the locations of the bands of interest if they are to be cut of the gel for further purification. - Put the gel slice into a 1.5 ml tube with 350 μl of RNA elution buffer.
- Incubate the tube overnight at 42 °C with 1,200 rpm shaking using Thermomixer C.
- Centrifuge the mixture at 4 °C for 10 min.
- Transfer the supernatant (about 300 μl) to a fresh tube.
- Add 100 μl of RNA elution buffer into the old tube with the gel slice, vortex and centrifuge for 5 min.
- Pool the supernatant into the supernatant previously collected (step A16).
- Add 1 μl of glycogen blue and 1 ml of 99.7% ice cold ethanol to the total supernatant.
- Mix and place the tube overnight at -20 °C or for 30 min at -80 °C.
- Centrifuge the tube at 16,000 x g for 15 min at 4 °C.
- Wash the blue pellet with 500 μl of 75% ice cold ethanol, then centrifuge at 12,000 x g for 5 min at 4 °C and remove the ethanol.
- Quick-spin the tube and carefully remove the residual ethanol.
- Air-dry the pellet for 5 min.
- Re-suspend the RNA pellet in 21 μl of DEPC H2O. Measure the concentration of RNA and calculate molarity. Normally, 1 μg 150 nt RNA approximately equals to 20 pmol.
- Adjust the final concentration of RNA sample to 5 μM (5 pmol/μl or 250 ng/μl).
- Store the RNA at -80 °C.
- Substrates of pri- and pre-miRNA are transcribed under the T7 promoter in vitro using PCR-generated templates. In this case, the forward PCR primer should contain T7 promoter sequences (TAATACGACTCACTATAG) to drive transcription directly from the PCR product.
- Removing 5' phosphates of pri- and pre-miRNAs
- Mix following in a fresh tube at 4 °C: 1 μl RNA (5 pmol), 2 μl of 10x NEB buffer 3, 2 μl of NEB Calf Intestine Alkaline Phosphatase (CIP), 1 μl of Superase-In (final concentration 1 U/μl), DEPC H2O 14 μl.
- Incubate at 37 °C for 30 min.
- Add 220 μl of DEPC H2O to the above reaction mixture.
- Add 240 μl of phenol: chloroform: isoamyl: alcohol (25:24:1) and vortex for 30 sec. Centrifuge for 5 min at RT and collect 210 μl aqueous phase.
- Add 210 μl chloroform: isoamyl alcohol (24:1) and vortex for 30 sec. Centrifuge for 5 min at RT and collect 170 μl aqueous phase.
- Add 510 μl of 99.7% ice cold ethanol, 17 μl 3 M NaAc (pH 5.2) and 1 μl of glycogen blue. Mix and leave the tube overnight at -20 °C or 30 min at -80 °C.
- Centrifuge at 16,000 x g for 15 min at 4 °C.
- Remove the supernatant carefully.
- Wash the blue pellet with 500 μl of 75% ice cold ethanol, then centrifuge at 12,000 x g for 5 min at 4 °C and remove the ethanol.
- Air-dry the pellet for 5 min.
- Re-suspend the RNA pellet in 4.5 μl of DEPC H2O.
- Mix following in a fresh tube at 4 °C: 1 μl RNA (5 pmol), 2 μl of 10x NEB buffer 3, 2 μl of NEB Calf Intestine Alkaline Phosphatase (CIP), 1 μl of Superase-In (final concentration 1 U/μl), DEPC H2O 14 μl.
- 5' end radioactive labeling of pri-and pre-miRNAs (this step must be performed in a radioisotope-designated space and with the appropriate safety training)
- Mix the following in a fresh tube at 4 °C: 4.5 μl of RNA (from step B), 1 μl of 10x NEB T4 kinase buffer, 3 μl of [γ-32P]-ATP (3,000 Ci/mmol 10 mCi/ml), 0.5 μl of Superase-In (final concentration 1 U/μl) and 1 μl of NEB T4 kinase.
- Incubate the reaction mixture at 37 °C for 2 h.
- Add 230 μl of DEPC H2O to the above reaction mixture.
- Add 240 μl of phenol: chloroform: isoamyl alcohol (25:24:1) and vortex for 30 sec. Centrifuge for 5 min at RT and take 210 μl aqueous phase.
- Add 140 μl of 5 M-ammonium acetate, 1 μl of glycogen blue and 1,050 μl of 99.7% ice cold ethanol. Mix and leave the tube overnight at -20 °C or 30 min at -80 °C.
- Centrifuge at 16,000 x g for 15 min at 4 °C.
- Remove the supernatant carefully.
- Wash the blue pellet with 500 μl of 75% ice cold ethanol, then centrifuge at 12,000 x g for 5 min at 4 °C and remove the ethanol.
- Air-dry the pellet for 5 min.
- Add 20 μl RNA loading buffer into the tube to dissolve the pellet. Here it can be stored at -20 °C.
- Prepare a 6% urea-polyacrylamide gel.
- Heat the tubes for 2 min at 95 °C. Cool down for 2 min on the ice.
- Briefly spin down the tubes.
- Flush the wells using the running buffer to remove the diffused urea.
- Load the RNA samples on 6% urea-PAGE gel and run at 350 V (20 V/cm) until bromophenol blue reaches the bottom of the gel.
- Remove one of the glass plates from the gel. Make sure to mark the position and orientation of the gel so that the gel can be aligned with the film once the film is developed. Wrap the gel with plastic wrap and place a phospho imager on the gel for 30 min. The radiolabeled transcript will appear as a strong band on the developed film.
- Align the photo with the gel and cut out the gel slice containing the labeled transcript. Put the gel slice in a 1.5 ml fresh tube with 350 μl of RNA elution buffer.
- Incubate the tube overnight at 42 °C with 1,200 rpm shaking using Thermomixer C.
- Transfer the supernatant (about 300 μl) to a fresh tube.
- Add 100 μl of RNA elution buffer into the old tube with the gel slice and vortex.
- Transfer the supernatant into the previous supernatant.
- Add 1 μl of glycogen blue and 1 ml of 99.7% ice cold ethanol into total supernatant.
- Mix and place the tube overnight at -20 °C or 30 min at -80 °C.
- Centrifuge the tube at 16,000 x g for 15 min at 4 °C.
- Wash the blue pellet with 500 μl of 75% ice cold ethanol, then centrifuge at 12,000 x g for 5 min at 4 °C and remove the ethanol.
- Quick-spin the tube and carefully remove the residual ethanol.
- Air-dry the pellet for 5 min. Be careful not to over-dry the pellet.
- Count cpm and re-suspend radiolabeled RNA in 20 μl RNA dissolving buffer. Denature the radiolabeled RNA in a 95 °C heating block for 2 min, and then take the entire 95 °C block out and fold the RNA by slowly cooling down the heat block to room temperature. Store the radiolabeled RNA at -20 °C.
- Mix the following in a fresh tube at 4 °C: 4.5 μl of RNA (from step B), 1 μl of 10x NEB T4 kinase buffer, 3 μl of [γ-32P]-ATP (3,000 Ci/mmol 10 mCi/ml), 0.5 μl of Superase-In (final concentration 1 U/μl) and 1 μl of NEB T4 kinase.
- Heterologous expression of Arabidopsis DCL1 complex in N. benthamiana
- Infiltrate N. benthamiana leaves with OD600 = 1 mixture of Agrobacterium tumefaciens ABI strains harboring pBA-2Flag-4Myc-DCL1, pBA-6Myc-HYL1, pBA-6Myc-SE, at OD600 of 0.8, 0.1, and 0.1 respectively. As a negative control we use A. tumefaciens ABI strains harboring an empty vector-pBA only.
- After two days, the infiltrated leaves are collected, ground in liquid nitrogen, and stored at -80 °C.
- Infiltrate N. benthamiana leaves with OD600 = 1 mixture of Agrobacterium tumefaciens ABI strains harboring pBA-2Flag-4Myc-DCL1, pBA-6Myc-HYL1, pBA-6Myc-SE, at OD600 of 0.8, 0.1, and 0.1 respectively. As a negative control we use A. tumefaciens ABI strains harboring an empty vector-pBA only.
- Immunoprecipitation of DCL1 microprocessor
- Make sure to cool down the centrifuges and rotors to 4 °C.
- Re-suspend the N. benthamiana powder sample in 5 volumes of the IP Buffer (typically 3 g of sample in 15 ml of IP buffer); keep on ice and protected from light (use aluminum foil to wrap the tube) until the sample is completely thawed. Transfer the solution to the 15 ml conical tubes.
- Centrifuge the samples at 16,000 x g for 15 min at 4 °C. Transfer all the supernatant into a new chilled 15 ml tube.
- Centrifuge at 4 °C for 15 min at 16,000 x g. Transfer all the supernatant into another new chilled 15 ml tube.
- During the centrifugation steps prepare anti-Flag antibody for immunoprecipitation. Add 400 μl bed volume Anti-FLAG® M2 magnetic beads (for 3 g plant tissue) into a new 2 ml Eppendorf tube on ice.
- Load the Eppendorf tube into the magnetic separator, DynaMagTM-2 to collect the beads. Remove the storage buffer quickly.
- Release the tube from DynaMagTM-2 and remove antibodies that are not conjugated to the DynaMag beads by incubation the Anti-FLAG® M2 magnetic beads with one bed volume of 0.1 M Glycine-HCl (pH 3.0).
- Invert the tube for 1.5 min (exactly).
- Load the Eppendorf tube into DynaMagTM-2, separate and remove the Glycine-HCl buffer quickly.
- Quickly equilibrate the Anti-FLAG® M2 magnetic beads with 1 ml TBS buffer three times using DynaMagTM-2.
- Wash the beads with 1 ml IP buffer for three times using DynaMagTM-2.
- Completely remove the IP buffer and add 400 μl new IP buffer into the tube to re-suspend the beads.
- Add the equilibrated beads into 15 ml tube of step E4.
- Then rotate the 15 ml tube at 4 °C for 2 h using a Rugged Rotator.
- While doing the first step immunoprecipitation with the Anti-FLAG® M2 magnetic beads, prepare anti-Myc antibody for the second-step immunoprecipitation.
- Add 120 μl bed volume of anti-c-myc agarose affinity gel into a new 1.5 ml tube. Then, wash the gel with 1 ml IP buffer.
- Spin the tube at 1,600 x g for 1min at 4 °C. Discard the supernatant.
- Repeat the washing for two more times.
- Take the majority of the washing buffer out and put anti-c-myc agarose affinity gel on ice.
- At the end of step E14, prepare 3x Flag peptide solution for elution of Flag-Myc-tagged DCL1 from the Anti-FLAG® M2 magnetic beads.
- Add 30 μl 3x Flag elution buffer stock (4 mg/ml) into 1.2 ml IP buffer to make a final concentration 100 μg/ml of 3x Flag peptide. Mix well and put it on ice.
- After step E14 is done, load the 15 ml tube (step E14) on the PolyATtract® System 1000 Magnetic Separation Stand for a few seconds, then slowly pour the supernatant to a trash can.
- Wash the wall of 15 ml conical tube with IP buffer until the supernatant is not green anymore, typically three times. For this, use the magnetic stand.
- Add 2 ml of IP buffer to the conical tube.
- Transfer all the beads carefully to a clean 2 ml Eppendorf tube.
- Load the 2 ml tube into DynaMagTM-2, and remove the IP buffer.
- Add 600 μl 3x Flag elution buffer into the 2 ml tube, incubate in rotation for 30 min at 4 °C.
- Load the 2 ml tube into DynaMagTM-2. Transfer the supernatant (1st elution solution) to the 1.5 ml Eppendorf tube containing anti-c-myc agarose affinity gel (step E19). Then, start second IP incubating the second tube in rotation at 4 °C.
- Add another 600 μl 3x Flag elution buffer to the anti-flag magnetic beads and perform a second round of elution for 30 min at 4 °C.
- Collect the second elution of the anti-Flag IP by placing the 2 ml Eppendorf tube into DynaMagTM-2. Pool this second elution with the first one in the 1.5 ml tube containing anti-c-myc agarose affinity gel (step E28).
- Continue the second IP step by rotating the 1.5 ml tube for 1.5 h at 4 °C.
- Centrifuge the 1.5 ml tube containing anti-c-myc agarose affinity gel (step E30) at 1,600 x g for 3 min at 4 °C. Remove the supernatant.
- Wash out the non-specific binding from the anti-c-myc agarose affinity gel by adding 1 ml IP buffer and inverting the tube 6-7 times. Centrifuge at 4 °C for 3 min at 1600 x g and remove the buffer.
- Re-suspend the affinity gel in 1 ml of IP buffer and aliquot it into eight new 1.5 ml siliconized tubes.
- Spin down the eight tubes at 1,600 x g for 3 min at 4 °C.
- Remove the buffer as complete as possible.
- Keep the eight tubes on ice for in vitro DCL1 reconstitution assay, as these purified DCL1 complexes are ready for in vitro assays.
Note: We prepare the fresh immunoprecipitate immediately before each assay.
- Make sure to cool down the centrifuges and rotors to 4 °C.
- In vitro DCL1 reconstitution assay (this step must be performed in a radioisotope-designated space and with the appropriate safety training)
- Carefully add 14 μl of fresh assay buffer and 1 μl radiolabeled pri-miRNA (approximately 2 x 103 cpm) into the eight tubes containing the immunoprecipitate. Gently re-suspend the agarose gel with the reaction mixture.
- Incubate the tube for 1 h at 37 °C with 900 rpm shaking using Thermomixer C.
- Gently mix the reaction every 5 min.
- After 1 h, add 170 μl of RNA elution buffer to stop assay.
- Add 200 μl of phenol: chloroform: isoamyl alcohol (25:24:1) and vortex for 30 sec.
- Centrifuge for 5 min at RT.
- Transfer 150 μl upper aqueous phase to the siliconized tubes.
- Add 20 μl of 3 M NaAc (pH 5.2), 1 µl of glycogen-blue, and 1 ml of 100% ice cold ethanol. Invert the tubes for several times and leave it overnight at -20 °C.
- Centrifuge at 16,000 x g for 15 min at 4 °C.
- Discard the supernatant carefully.
- Wash the blue pellet with 500 μl of 75% ice cold ethanol, then centrifuge at 12,000 x g for 5 min at 4 °C and remove the ethanol.
- Quick spin down the tubes, remove the residue ethanol using pipette.
- Air-dry the pellet for 5 min.
- Suspend pellets with 20 µl RNA loading buffer. The sample can now be stored at -20 °C.
- Boil the tubes for 1 min at 95 °C. Cool down for 2 min on the ice.
- Briefly spin down the tubes.
- Prepare a 15% urea-polyacrylamide gels, and flush the wells of gel to remove the diffused urea before loading the samples.
- Load the assay samples and RNA marker on 15% urea-polyacrylamide gels. The RNA marker should be 5′-end labeled according to the manufacturer’s protocol with [γ-32P]-ATP.
- Run at 350 V (20 V/cm) for approximately one hour until bromophenol blue reaches the bottom of the gel.
- Disassemble the gel cast.
- Place the gel into a tank containing fixing buffer for 30 min.
- Dry the gel with a Gel Dryer under the gradient cycle at 80 °C for 2-4 h.
- Cover the dried gel with plastic wrap and place a phosphate screen on the gel for 2 h or overnight.
- The processed RNA products will appear as a strong band on phospho imager (PharosFXTM Plus System) and quantified with Quantity One Version 4.6.9 according to the manufacturer’s instructions.
- Carefully add 14 μl of fresh assay buffer and 1 μl radiolabeled pri-miRNA (approximately 2 x 103 cpm) into the eight tubes containing the immunoprecipitate. Gently re-suspend the agarose gel with the reaction mixture.
Representative data
Representative data could refer to the Figures 2, 3, 4, 5, 6, 7 and 8 from Zhu et al. (2013), as well as supplementary Figures 2, 3, 4, 5, 6 and 9.
Notes
- The E and F steps should be performed successively in one day.
- The C and F steps must be performed in a radioisotope-designated space and with the appropriate safety training.
Recipes
- 5x transcription buffer
400 mM HEPES (pH 7.5)
10 mM spermidine
200 mM DTT
125 mM MgCl2 and 20 mM of each NTP - RNA loading buffer
95% deionized formamide
0.025% bromophenol blue
0.025% xylene cyanol
5 mM EDTA and 0.025% SDS - RNA elution buffer
0.3 M NaAc (pH 5.2) and 2% SDS - DEPC H2O
Add 1 ml of DEPC to 1 L of H2O, shake vigorously and autoclave. - RNA dissolve buffer
100 mM KCl
30 mM Tris-HCl (pH 7.5) - Chloroform: isoamyl alcohol (24:1)
Chloroform 96 ml
Isoamyl alcohol 4 ml - IP buffer
40 mM Tris-HCl (pH 7.5)
300 mM KCl
5 mM MgCl2
5 mM DTT
0.2 mM EDTA (pH 8.0)
0.2% Triton X-100
1 mM PMSF
2% glycerol
0.3% (vol/vol) protease inhibitor cocktail stock
1 tablet per 25 ml IP buffer of EDTA-free protease inhibitor cocktail - Protease inhibitor cocktail stock
Protease inhibitor cocktail (about 89 mg) is dissolved in 10 ml ddH2O - TBS buffer
50 mM Tris HCl (pH 7.4)
150 mM NaCl - 3x Flag elution buffer stock (4 mg/ml)
Add 4 mg 3x Flag peptide into 1 ml IP buffer - Washing buffer
20 mM Tris-HCl (pH 7.5)
1 mM DTT
4 mM MgCl2 and 100 mM KCl - Assay buffer
20 mM Tris-HCl (pH 7.5)
4 mM MgCl2
1 mM DTT
10 mM ATP
2 mM GTP
2 U/μl Supernase-In - Fixing buffer
40% ethanol
10% acetic acid and 5% glycerol
Acknowledgments
This work was supported by grants from the US National Science Foundation (NSF) CAREER (MCB-1253369), the US National Institutes of Health (R21AI097570) and the Welch foundation (A-1777) to X.Z, and also supported by Chinese Universities Scientific Fund (2014RC006) to H.Z.
References
- Han, J., Lee, Y., Yeom, K. H., Kim, Y. K., Jin, H. and Kim, V. N. (2004). The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 18(24): 3016-3027.
- Zhang, X., Henriques, R., Lin, S. S., Niu, Q. W. and Chua, N. H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1(2): 641-646.
- Zhu, H., Zhou, Y., Castillo-Gonzalez, C., Lu, A., Ge, C., Zhao, Y. T., Duan, L., Li, Z., Axtell, M. J., Wang, X. J. and Zhang, X. (2013). Bidirectional processing of pri-miRNAs with branched terminal loops by Arabidopsis Dicer-like1. Nat Struct Mol Biol 20(9): 1106-1115.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Wang, T., Castillo-González, C., You, L., Li, R., Wen, L., Zhu, H. and Zhang, X. (2015). In vitro Reconstitution Assay of miRNA Biogenesis by Arabidopsis DCL1. Bio-protocol 5(8): e1454. DOI: 10.21769/BioProtoc.1454.
Category
Plant Science > Plant molecular biology > RNA > RNA interference
Plant Science > Plant biochemistry > Protein > Isolation and purification
Molecular Biology > RNA > RNA interference
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









