- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Construction of Glycine Oxidase Mutant Libraries by Random Mutagenesis, Site Directed Mutagenesis and DNA Shuffling
Published: Vol 4, Iss 19, Oct 5, 2014 DOI: 10.21769/BioProtoc.1252 Views: 10987
Reviewed by: Kanika GeraAksiniya AsenovaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Novel Method of Inducible Directed Evolution to Evolve Complex Phenotypes
Ibrahim S. Al’Abri [...] Nathan Crook
Oct 20, 2022 3062 Views

Comprehensive Mapping of EZ-Tn5 Transposon Insertion Sites in Pseudomonas argentinensis SA190 Using RATE-PCR
Büsra Elkatmis [...] Maged M. Saad
Jul 20, 2025 1751 Views

Editing the Serratia proteamaculans Genome Using the Allelic Exchange Method
Ksenia Chukhontseva [...] Ilya Demidyuk
Sep 20, 2025 1387 Views
Abstract
Glyphosate, a broad spectrum herbicide widely used in agriculture all over the world, inhibits 5-enolpyruvylshikimate-3-phosphate synthase in the shikimate pathway, and glycine oxidase (GO) has been reported to be able to catalyze the oxidative deamination of various amines and cleave the C-N bond in glyphosate (Pedotti et al., 2009). Here, in an effort to improve the catalytic activity of the glycine oxidase that was cloned from a glyphosate-degrading marine strain of Bacillus cereus (BceGO), we used a bacteriophage T7 lysis-based method for high-throughput screening of oxidase activity and engineered the gene encoding BceGO by directed evolution.
Materials and Reagents
- Bacillus cereus HYC-7
- Escherichia coli (E.coli) DH5α strain, bacteriophage T7
- Glyphosate (Sigma-Aldrich, catalog number: PS1051 )
- Tryptone (Difco)
- Yeast extract (Difco)
- Ampicillin
- o-Dianisidine dihydrochloride (Sigma-Aldrich, catalog number: D3252 )
- Horseradish peroxidase (Sigma-Aldrich, catalog number: P6782 )
- Protein expression vector of pGEX-6P-1 (the plasmid full length 4,984 bp) (GE Healthcare, catalog number: 28-9546-48 ; Genbank accession number: U78872.1)
- Recombinant plasmid pGEX-GO contains encoding gene of glycine oxidase from Bacillus cereus HYC-7
The nucleotide sequence (1,110 bp) was submitted to the NCBI Genbank and gained the accession number (KC203486.1). - Taq DNA polymerase (Takara, catalog number: R500A )
- DpnI restriction enzyme (Takara, catalog number: 1235A )
- dATP, dTTP, dCTP, dGTP (Takara, catalog numbers: 4026Q , 4029Q , 4028Q , 4027Q )
- TransStart® FastPfu DNA polymerase (TransGen Biotech, catalog number: AP221-01 )
- High Pure dNTPs (TransGen Biotech, catalog number: AD101-01 )
- Luria-Bertani medium (see Recipes)
Equipment
- 96 deep-well plates (Axygen, catalog number: P-DW-20-C-S )
- Gel purification column (Axygen)
- Thermo Multiskan spectrum plate reader (Thermo Scientific, catalog number: 51118600 )
- Thermal cyclers (Bio-Rad Laboratories, catalog number: 186-1096 )
- Ultrasonic processor (Sigma-Aldrich, catalog number: Z412619-1EA )
Procedure
- Random mutagenesis
- Prepare the amplification mixture (100 µl) as follows:
10 µl of 10x Taq buffer (Mg2+ plus)
5 µl of 10 mM Mn2+
2 µl of 10 mM dGTP and dCTP
1 µl of 10 mM dATP and dTTP
2 µl of 100 nM oligonucleotide primer F
2 µl of 100 nM oligonucleotide primer R
1 µl of recombinant plasmid pGEX-GO as template
2 µl of Taq DNA polymerase
Add ddH2O to a final volume of 100 μl - The error-prone PCR procedure was performed using the following parameters:
Segment Cycles Temperature Time 1 1 94 °C 3 min 2 30 94 °C 30 sec 59 °C 30 sec 72 °C 80 sec 3 1 72 °C 7 min - Check product by electrophoresis of 5 μl of error-prone PCR product on 1% agarose gel.
- Error-prone PCR products were purified, digested with BamHI and XhoI, cloned into pGEX-6P-1, and transformed into E.coli DH5α to construct the random mutant library.
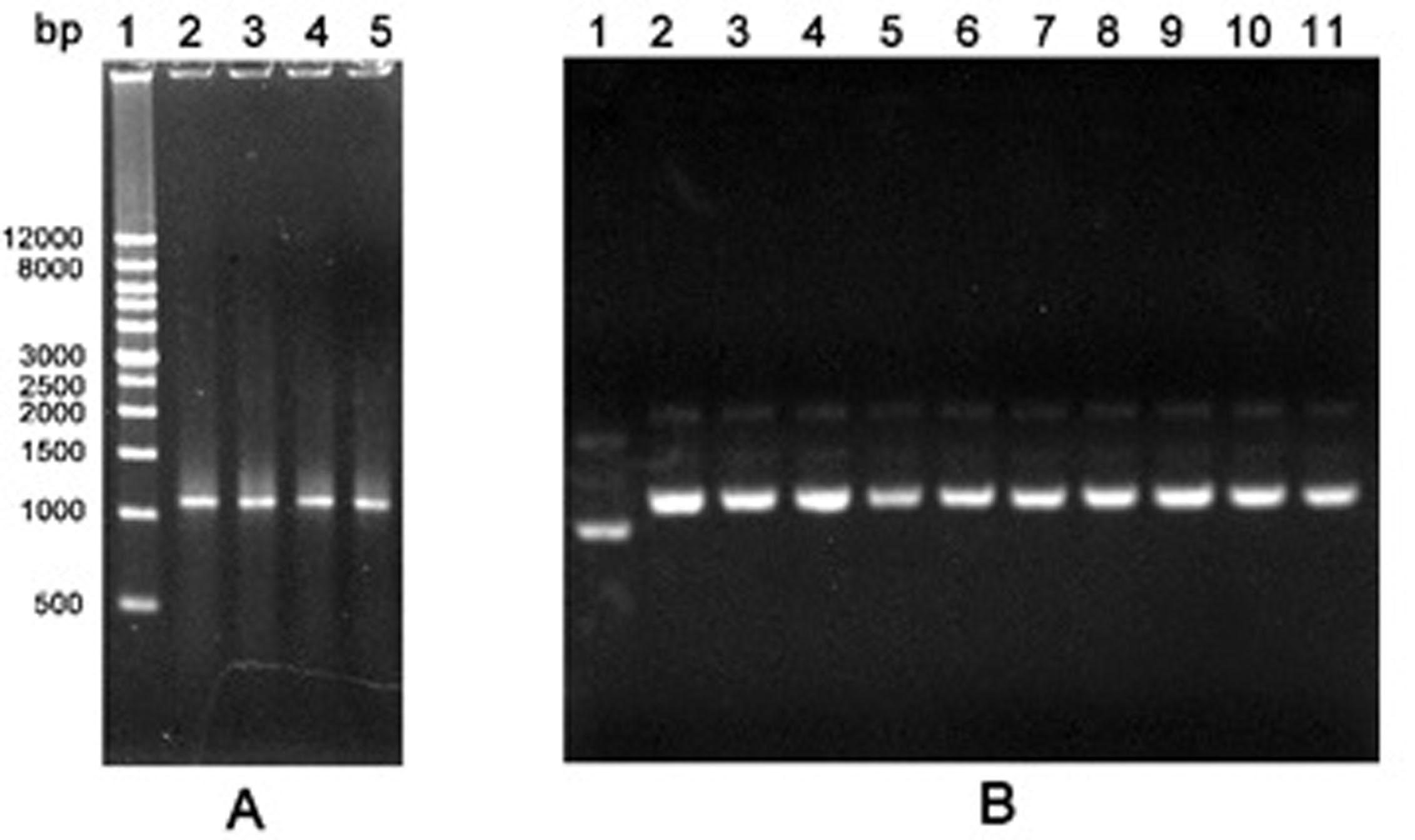
Figure 1. Agarose gel electrophoresis of PCR products by the first round error-prone PCR and recombinant plasmids. A. PCR products. Lane 1: Wide Range DNA Marker (500~12,000 bp); Lane 2-5: Error-prone PCR products; B. Recombinant plasmids. Lane 1: The empty vector pGEX-6P-1; Lane 2-11: Recombinant plasmids pGEX-GOs from colonies.
- Prepare the amplification mixture (100 µl) as follows:
- Site directed mutagenesis
- Prepare the amplification mixture (50 µl) as follows:
10 µl of 10x FastPfu buffer (Mg2+ plus)
1 µl of 10 mM high pure dNTPs
1 µl of 100 nM oligonucleotide primer F
1 µl of 100 nM oligonucleotide primer R
1 µl of dsDNA template
1 µl of FastPfu DNA polymerase
Add ddH2O to a final volume of 50 μl - The site directed mutagenesis PCR procedure was performed as the following parameters:
Segment Cycles Temperature Time 1 1 97 °C 2 min 2 20 94 °C 20 sec 54 °C 30 sec 72 °C 1 min/kb of plasmid length 3 1 72 °C 7 min - Check product by electrophoresis of 5 μl of the site directed mutagenesis PCR product on 1% agarose gel.
- The site directed mutagenesis PCR products were purified, digested with DpnI, and transformed into E.coli DH5α.
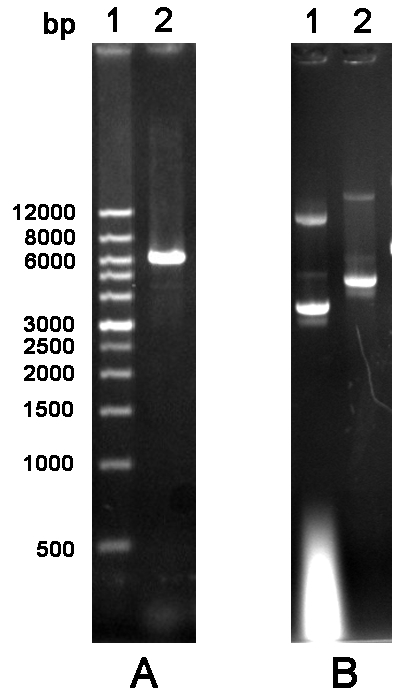
Figure 2 Agarose gel electrophoresis analysis of cylcled PCR product and mutated plasmid. A. PCR product. Lane 1: Wide Range DNA Marker (500~12,000 bp); Lane 2: Cycled PCR product. B. Mutated plasmid. Lane 1: The empty vector pGEX-6P-1; Lane 2: Mutated plasmid.
- Prepare the amplification mixture (50 µl) as follows:
- DNA shuffling
- Obtaining DNA fragments for shuffling.
- Prepare the parental genes by PCR amplification (100 µl) as follows:
10 µl of 10x Taq buffer (Mg2+ plus)
2 µl of 10 mM High Pure dNTPs
2 µl of 100 nM oligonucleotide nested primer F1
2 µl of 100 nM oligonucleotide nested primer R1
1 µl of beneficial mutatant as template
2 µl of Taq DNA polymerase
Add ddH2O to a final volume of 100 μl - The DNA shuffling PCR procedure was performed as the following parameters:
Segment Cycles Temperature Time 1 1 94 °C 3 min 2 30 94 °C 30 sec 62 °C 30 sec 72 °C 80 sec 3 1 72 °C 7 min - Check product by electrophoresis of 5 μl of product on 1% agarose gel.
- Mix ~1 μg of each purified PCR products.
- Prepared for DNA fragmentation by ultrasonic treatment at 0 °C for 40 min to generate a pool of fragments, then the DNA fragments between 100~200 bp were purified using gel purification column.
- Check the DNA fragments by electrophoresis of 10 μl of product on 3% agarose gel.

Figure 3. Schematic of parental genes with the nested primers in the process of DNA shuffling
- Prepare the parental genes by PCR amplification (100 µl) as follows:
- Reassembled by primerless PCR.
- Prepare the amplification mixture (50 µl) as follows:
10 µl of 10x Taq buffer (Mg2+ plus)
2 µl of 10 mM high pure dNTPs
42 µl of purified fragment DNA
1 µl of Taq DNA polymerase
Add ddH2O to a final volume of 50 μl - The primerless PCR procedure was performed as the following parameters:
Segment Cycles Temperature Time 1 1 94 °C 3 min 2 60 94 °C 30 sec 40 °C 30 sec 72 °C 20 sec + 1 sec per cycle 3 1 72 °C 7 min - Check product by electrophoresis of 5 μl of PCR product on 1% agarose gel. A smear of reassembled product that extends above the molecular weight of the parent gene should be visible.
- Prepare the amplification mixture (50 µl) as follows:
- Amplification of full-length sequences.
- Prepare the amplification mixture (50 µl) as follows:
10 µl of 10x Taq buffer (Mg2+ plus)
2 µl of 10 mM high pure dNTPs
1 µl of 100 nM oligonucleotide nested primer F2
1 µl of 100 nM oligonucleotide nested primer R2
5 µl of unpurified reassembly reaction mixture as template
1 µl of Taq DNA polymerase
Add ddH2O to a final volume of 50 μl - The PCR procedure was performed as the following parameters:
Segment Cycles Temperature Time 1 1 94 °C 3 min 2 30 94 °C 30 sec 59 °C 30 sec 72 °C 80 sec 3 1 72 °C 7 min - Check product by electrophoresis of 5 μl of product on 1% agarose gel.
- PCR products were purified, digested with BamHI and XhoI, cloned into pGEX-6P-1, and transformed into E.coli DH5α to create the DNA shuffling library.
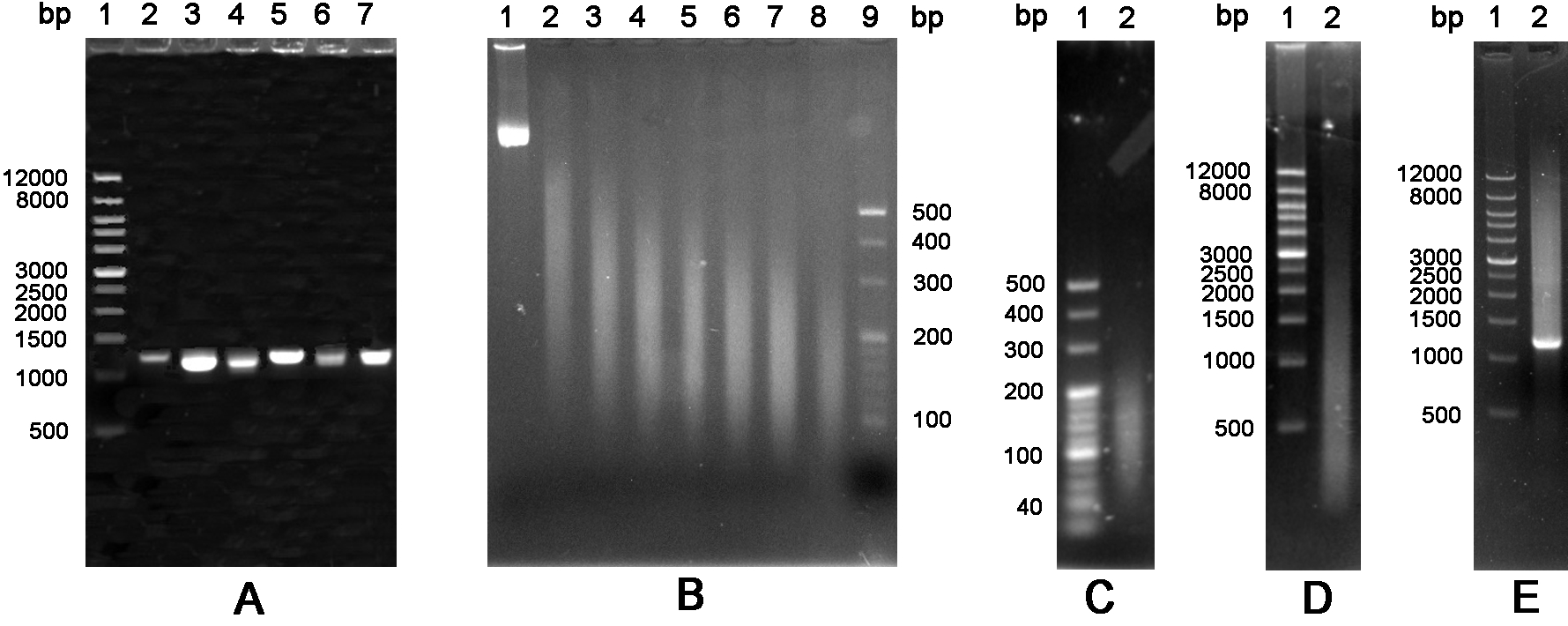
Figure 4. The schematic of glyphosate oxidase gene by DNA shuffling. A. Amplification of six variants GO. Lane 1: Wide Range DNA Marker (500~12,000 bp); Lanes 2-7: DNA fragments encoding variants GO. B. DNA fragmentation. Lanes 1-8: DNA fragments were treated by ultrasonic at 0 °C with a time gradient of 5 min from 0 to 35 min; Lane 9: 20 bp DNA Ladder Marker (20~500 bp). C. Purification of DNA fragments. Lane 1: 20 bp DNA Ladder Marker (20~500 bp); Lane 2: DNA fragments of 100~200 bp were purified from an agarose gel. D. Fragments were reassembled without primers. Lane 1: Wide Range DNA Marker (500~12,000 bp); Lane 2: DNA fragments were reassembled into a full-length gene by 60 cycles without primers. E. The reassembled full-length products were amplified by the standard PCR. Lane 1: Wide Range DNA Marker (500~12,000 bp); Lane 2: PCR product with primers.
- Prepare the amplification mixture (50 µl) as follows:
- Obtaining DNA fragments for shuffling.
- Screening
- The resulting library of BceGO mutants were expressed into 96 deep-well plates (containing 0.6 ml Luria-Bertani medium) and transferred onto Luria-Bertani agar plates as corresponding copies, followed by an overnight growth (37 °C, 300 rpm).
- When the cultures grew to saturation, both IPTG (at a final concentration of 0.1 mM) and the bacteriophage T7 (above 100 particles per cell) were added into 96 deep-well plates to synchronize the induction of recombinant mutants with the release of the lysis of the host E.coli DH5α at 37 °C with shaking for 6 h.
- The enzyme-coupled colorimetric assay (200 µl) was performed as follows:
159 µl of lysis cell extracts
20 µl of 50 mM glyphosate (at a decreasing substrate concentration gradient in sequential rounds of screening system)
20 µl of 0.32 mg/ml o-dianisidine dihydrochloride
1 µl of 5 unit/ml horseradish peroxidase
Then incubated at 25 °C for 8 h - The absorbance change at 450 nm for each well in the microtiter plates was measured and compared with the control (harboring wild-type BceGO or containing the empty vector pGEX-6P-1). Mutants that outperformed the wild-type were chosen for further activity analysis (Figure 1).
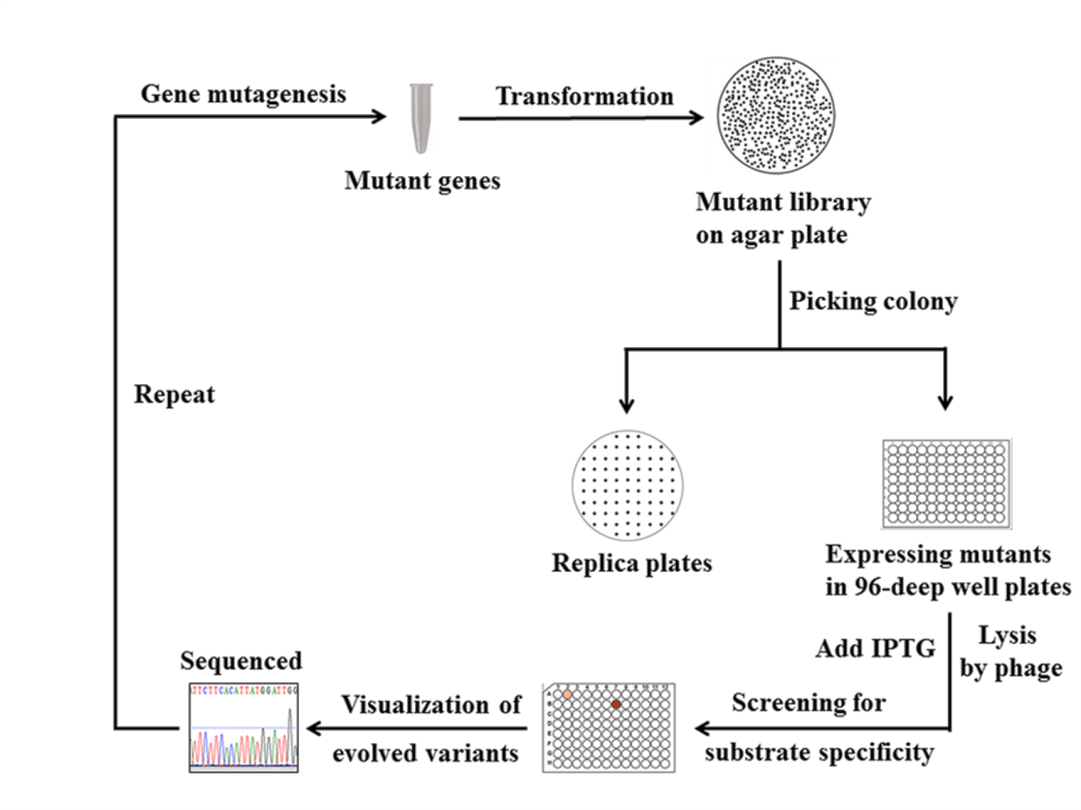
Figure 5. The screening process of glycine oxidase mutant library
- The resulting library of BceGO mutants were expressed into 96 deep-well plates (containing 0.6 ml Luria-Bertani medium) and transferred onto Luria-Bertani agar plates as corresponding copies, followed by an overnight growth (37 °C, 300 rpm).
Representative data
Table 1. The apparent kinetic parameters on glycine and glyphosate measured for wild-type BceGO and variants obtained by random mutagenesis, site saturation mutagenesis and DNA shuffling
| Glycine | Glyphosate | |||
| kcat,app (min-1) | Km,app (mM) | kcat,app (min-1) | Km,app (mM) | |
| Wild-type | 8.17 ± 0.31 | 1.04 ± 0.17 | 5.72 ± 0.42 | 84.79 ± 4.25 |
| 22D11 | 1.16 ± 0.05 | 54.6 ± 3.47 | 2.95 ± 0.21 | 8.29 ± 0.27 |
| 23B1 | 4.56 ± 0.38 | 0.99 ± 0.04 | 7.15 ± 0.62 | 18.88 ± 2.52 |
| B1R | 0.44 ± 0.03 | 58.5 ± 5. 26 | 2.78 ± 0.46 | 2.45 ± 0.15 |
| B2R11 | 1.86 ± 0.09 | 105.6 ± 7.31 | 3.83 ± 0.17 | 2.77 ± 0.21 |
| B2R14 | 1.35 ± 0.12 | 92.5 ± 7. 43 | 4.16 ± 0.14 | 2.17 ± 0.37 |
| B2R23 | 13.02 ± 0.96 | 101.8 ± 8.29 | 30.80 ± 1.33 | 3.80 ± 0.26 |
| B2R81 | 5.41 ± 0.83 | 134.4 ± 10.33 | 7.27 ± 0.75 | 4.37 ± 0.30 |
| B3S1 | 5.43 ± 0.79 | 41.55 ± 3.32 | 11.67 ± 0.98 | 0.53 ± 0.03 |
| B3S4 | 5.68 ± 0.64 | 80.43 ± 5.01 | 12.99 ± 1.14 | 1.37 ± 0.07 |
| B3S6 | 10.14 ± 1.32 | 138.1 ± 12.16 | 13.22 ± 1.78 | 1.69 ± 0.08 |
| B3S7 | 2.30 ± 0.31 | 41.64 ± 2.10 | 4.63 ± 0.39 | 0.57 ± 0.02 |
Especially, B3S1 demonstrated a 160-fold increase in substrate affinity for glyphosate, a 326-fold increase in catalytic efficiency towards glyphosate and a significant enhancement in the specificity constant over the wild-type BceGO, achieving the goal of efficient oxidation of glyphosate by evolution of glycine oxidase.
Notes
- The error-rate is controlled by varying the amount of MnCl2 and unblanced dNTPs added to the error-prone PCR reaction.
- Design nested primer to amplify the parental genes for the DNA fragmentation and full-length sequences from reassembly products.
Recipes
- Luria-Bertani medium
10 g/L tryptone
5 g/L yeast extract
10 g/L NaCl
Ampicillin (50 mg/L) was added as needed.
Acknowledgments
We thank Drs. Ziduo Liu and Dexin Kong for valuable discussions about this article.
References
- GST Gene Fusion System Handbook. (2002). Biosciences, Amersham.
- Zhan, T., Zhang, K., Chen, Y., Lin, Y., Wu, G., Zhang, L., Yao, P., Shao, Z. and Liu, Z. (2013). Improving glyphosate oxidation activity of glycine oxidase from Bacillus cereus by directed evolution. PLoS One 8(11): e79175.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Zhan, T. (2014). Construction of Glycine Oxidase Mutant Libraries by Random Mutagenesis, Site Directed Mutagenesis and DNA Shuffling. Bio-protocol 4(19): e1252. DOI: 10.21769/BioProtoc.1252.
Category
Microbiology > Microbial genetics > Mutagenesis
Molecular Biology > DNA > Mutagenesis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










