- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Metabolite and Fatty Acid Analysis of Yeast Cells and Culture Supernatants
Published: Vol 4, Iss 17, Sep 5, 2014 DOI: 10.21769/BioProtoc.1219 Views: 17849
Reviewed by: Claudia CatalanottiAksiniya AsenovaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Sterol Analysis in Kluyveromyces lactis
Yvetta Gbelska [...] Alexandra Konecna
Sep 5, 2017 7788 Views

Tracking Lipid Transfer by Fatty Acid Isotopolog Profiling from Host Plants to Arbuscular Mycorrhiza Fungi
Andreas Keymer [...] Caroline Gutjahr
Apr 5, 2018 9254 Views

Integrated Membrane Yeast Two-Hybrid System for the Analysis of Membrane Protein Complexes
Brianna L. Greenwood [...] David T. Stuart
Aug 20, 2025 3020 Views
Abstract
Metabolite and fatty acid analysis play important roles in evaluating the metabolic state of microorganisms. To examine the growth state and metabolism response of cells to environmental stress or genetic modification, intracellular and extracellular metabolites including fatty acids are usually analyzed to help understand the cellular biochemical changes in microorganisms. In this protocol, gas chromatography-mass spectrometry based analysis was employed to investigate the fatty acids and other metabolites in yeast cells.
Keywords: Metabolite profilingMaterials and Reagents
- Saccharomyces cerevisiae (S. cerevisiae) cells (EUROSCARF)
- Methanol (Sigma-Aldrich, catalog number: 322415 )
- Ethanol (Sigma-Aldrich, catalog number: 277649 )
- Chloroform (Sigma-Aldrich, catalog number: C2432 )
- Acetic acid (Sigma-Aldrich, catalog number: 320099 )
- Heptadecanoic acid (Sigma-Aldrich, catalog number: H3500 )
- Heptanoic acid (Sigma-Aldrich, catalog number: 75190 )
- Ribitol (Sigma-Aldrich, catalog number: A5502 )
- Fatty acid methyl esters (FAMEs) mix C8-C24 (Sigma-Aldrich, Supelco, catalog number: 18918 )
- 10% BF3-methanol (Fluka, catalog number: 15716 )
- Hexane (Sigma-Aldrich, catalog number: 34859 )
- SPE column (200 mg/3 ml) (Phenomenex, Strata, model: C18-E )
- Adapter (Sigma-Aldrich, Supelco, catalog number: 57020-U )
- Disposable syringe (Sigma-Aldrich, catalog number: Z116866 )
- Sodium chloride (Sigma-Aldrich, catalog number: 746398 )
- Methoxyamine hydrochloride (Sigma-Aldrich, catalog number: 226904 )
- N-methyl-N-(trimethylsilyl)-trifluoroacetamide (MSTFA) with 1% trimethylchlorosilane (TMCS) (Fluka, catalog number: 69478 )
- Agilent Technologies Vial Scr Fix Insert Clr PK100 (Agilent, catalog number: 5188-6591 )
- 10 mg/ml heptadecanoic acid (see Recipes)
- 10 mg/ml heptanoic acid (see Recipes)
- 2 mg/ml ribitol (see Recipes)
- Saturated NaCl solution (see Recipes)
Equipment
- Glass beads (acid-washed, 425-600 µm) (Sigma-Aldrich, catalog number: G8772 )
- Fume hood
- SSI 2341-S0S screw cap tubes
- 10 ml PP conical bottom tubes
- HP-5MS capillary column (30 m x 0.250 mm i.d.; film thickness: 0.25 µm) (Agilent)
- Centrifugal Evaporator RC10.09 (Thermo Fisher Scientific, catalog number: 11176780 )
- Thermostatic Cabinet Lovibond ET619-4 (Tintometer GmbH, catalog number: ET2428210 )
- Vortex mixer (CabNet)
- Thermo-shaker
- Eppendorf centriguge 5810R (10,000 rpm) (Eppendorf)
- Thermo scientific Sorvall Legend Micro 21R centrifuge (12,000 rpm-14,000 rpm) (Thermo Fisher Scientific)
- 30 °C Orbital shaker incubator (Yihder Technology, model: LM-570RD )
- NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific)
- FastPrep® -24 instrument (MP Biomedicals, model: 6004-500 )
- Gas chromatography-mass spectrometry (GC-MS) system (Agilent, model: 7890A-5975C ) with an autoinjector G4513A
Software
- Agilent MSD Chemstation data analysis software
Procedure
Note: Workflow of metabolite and fatty acid analyses of yeast cells and culture supernatants is shown in Figure 1.
- Sample preparation for metabolite and fatty acid analysis
- Samples of the yeast culture were collected at different time points and the OD600 was measured using a NanoDrop 2000c spectrophotometer.
Note: For Saccharomyces cerevisiae cells, OD600 of 1 is roughly equal to 1 x 107 cells/ml. When the OD of cells cultured in medium containing fatty acid is determined, the cells should be washed by 0.9% NaCl solution at least 3 times before OD measurement, since the fatty acid affects the spectral absorbance. If the cells are cultured in YNB or YPD medium, no washing step is needed. Usually, S. cerevisiae cells were cultured at 30 °C. - After centrifugation for 10 min at 10,000 rpm, 4 °C, the supernatant was transferred into a fresh tube by a micropipettor. The cell pellet was resuspended in 0.9% NaCl solution to same OD600.
- An equal number of yeast cells (8 x 107) were collected by centrifugation from different yeast cultures and/or time points for metabolite and fatty acid extraction (proceed to step A4). An equal volume of cell culture supernatant (10 ml) was collected for extracellular fatty acid analysis (proceed to step A5).
- The cell pellet collected in step A3 was washed twice with cold methanol (< -40 °C) and then collected. The intracellular metabolites and lipids were extracted from control and experimental cells using a chloroform:methanol (2: 1) method adapted from Browse et al. (1986).
- The cell pellet was resuspended in 1 ml of 0.9% NaCl solution and then acidified with 200 µl of acetic acid.
- As internal standard (IS) to correct for metabolite loss during sample preparation, 10 µl of 10 mg/ml heptadecanoic acid and heptanoic acid dissolved in ethanol and 10 μl of 2 mg/ml ribitol dissolved in water were added to the extraction solvent.
- After adding approximately 300 µl of glass beads, the cells were disrupted in a FastPrep®-24 instrument for 30 sec and cooled in ice for 30 sec; this procedure was repeated 4 times.
- 3 ml of a chloroform-methanol 2: 1 mixture was added, and the samples were inverted several times, vortexed vigorously, and centrifuged at 10,000 x g for 10 min at 4 °C. The aqueous layer (upper) and cell debris (middle layer) were transferred to a new tube using a micropipetor.
- Chloroform layer (lower) was collected. An additional 2 ml of the chloroform was added to the aqueous and cell debris layer to further extract lipids. The generated chloroform layer (lower) was also collected.
- The chloroform layer was combined and rotary evaporated to dryness using a Centrifugal Evaporator at 30 °C.
- For metabolite profiling, 1 ml of the aqueous phase layer (generated in step A4e) was collected and evaporated at 30 °C overnight to dryness too. The residual aqueous layer and cell debris was discarded.
Note: Samples need to be completely dry otherwise the remaining water could inhibit the following reaction.
- The cell pellet was resuspended in 1 ml of 0.9% NaCl solution and then acidified with 200 µl of acetic acid.
- For the fatty acids in the culture medium, the Solid-Phase Extraction (SPE) method (Horak et al., 2009) was adopted.
- First, 0.5 ml 1 M HCl, 2 µl 10 mg/ml heptadecanoic acid and 2 µl 10 mg/ml heptanoic acid were added into 10 ml culture supernatant and mixed.
- Next, the SPE column was activated with 2.5 ml methanol and washed with 5 ml water. After that, the 10 ml culture supernatant in step A5a was passed through the SPE column.
Note: Syringe and adapter were used to load liquid onto the SPE column. - The SPE column was washed with 5 ml water and dried at room temperature.
Note: The SPE column must be dried. Otherwise the reproducibility will be affected. - 1.5 ml chloroform was loaded onto the SPE column for fatty acid elution. The effluent chloroform from the column was collected and also evaporated to dryness. It can be evaporated in a rotary evaporator at 30 °C for several hours or by leaving the tube open in a fume hood overnight.
- First, 0.5 ml 1 M HCl, 2 µl 10 mg/ml heptadecanoic acid and 2 µl 10 mg/ml heptanoic acid were added into 10 ml culture supernatant and mixed.
- Samples of the yeast culture were collected at different time points and the OD600 was measured using a NanoDrop 2000c spectrophotometer.
- Fatty acid analysis
- Fatty acid derivatization was performed according to previous work (Horak et al., 2009). Fatty acid methyl esters (FAMEs) mix C8-C24 was used as a standard.
- The dried lipid residue from step A4f or A5d was redissolved in 500 µl 10% BF3-methanol and incubated in a sealed screw cap tube in a 95 °C heat block for 20 min.
- Cool down the tube to near room temperature. 300 µl saturated NaCl in water was added into the reaction, and FAMEs were extracted from the mixture with 300 µl n-hexane. Samples were centrifuged at 14,000 rpm for 1 h at room temperature. The upper hexane layer was next transferred to glass vials for GC-MS analysis.
- The dried lipid residue from step A4f or A5d was redissolved in 500 µl 10% BF3-methanol and incubated in a sealed screw cap tube in a 95 °C heat block for 20 min.
- Chromatography was performed using Agilent Technologies GC-MS system equipped with a HP-5MS capillary column.
- Samples of 1 µl were injected into the column by splitless mode using an autoinjector. Helium was used as a carrier gas at 1.1 ml/min. The inlets and MS source temperatures were maintained at 250 and 230 °C, respectively. The oven temperature was maintained at 80 °C for 1 min and ramped to 250 °C at a rate of 7 °C/min, then held at 250 °C for 10 min. Data were acquired in a full scan from 35 to 600 m/z. The representative GC-MS spectra for yeast fatty acid profile is shown in Figure 2.
- The detected FAME peaks were integrated after noise reduction and baseline correction. FAME standard curves were made by running different concentrations (0.125-1 mg/ml) of FAMEs mix C8-C24 and IS. The amount of fatty acids were calculated based on extrapolation for the IS and the relative response factors were also calculated. A representative standard curve of C16: 1 m.e. is shown in Figure 3.
- Samples of 1 µl were injected into the column by splitless mode using an autoinjector. Helium was used as a carrier gas at 1.1 ml/min. The inlets and MS source temperatures were maintained at 250 and 230 °C, respectively. The oven temperature was maintained at 80 °C for 1 min and ramped to 250 °C at a rate of 7 °C/min, then held at 250 °C for 10 min. Data were acquired in a full scan from 35 to 600 m/z. The representative GC-MS spectra for yeast fatty acid profile is shown in Figure 2.
- Fatty acid derivatization was performed according to previous work (Horak et al., 2009). Fatty acid methyl esters (FAMEs) mix C8-C24 was used as a standard.
- Metabolic profiling
- The sample for metabolic profiling was derivatized according to previous work (Wang et al., 2010).
- The dry residue from step A4g was re-dissolved in 50 µl of 20 mg/ml solution of methoxyamine hydrochloride in pyridine and kept at 37 °C for 60 min for carbonyls protection.
- 100 µl of N-methyl-N-(trimethylsilyl)-trifluoroacetamide (MSTFA) with 1% trimethylchlorosilane (TMCS) was further added to each sample, and silylation was carried out at 70 °C for 30 min.
- After incubation, samples were centrifuged at 14,000 rpm for 1 h at room temperature and then transferred to glass vials for GC-MS analysis.
Note: It would be better that all samples were analyzed within 12 h after they are obtained. The samples can be stored in the sealed vials for a maximum of 2 days at -20 °C. Because the derivative metabolites might be unstable after long term storage, the concentration might decrease.
- The dry residue from step A4g was re-dissolved in 50 µl of 20 mg/ml solution of methoxyamine hydrochloride in pyridine and kept at 37 °C for 60 min for carbonyls protection.
- The same GC-MS system and column (as indicated in step B2) were used.
- Samples of 1 µl were injected into the column by splitless mode using an autoinjector. Helium was used as a carrier gas at 1.1 ml/min. The inlets and MS source temperatures were maintained at 250 and 230 °C, respectively. The oven temperature was maintained at 75 °C for 4 min and raised at 4 °C/min to a final temperature of 280 °C and held for 2 min. Mass spectra were recorded from 35 to 600 m/z.
- Chromatogram acquisition and mass spectra identification were obtained using the Agilent MSD Chemstation Data Analysis software. Chemical identification of the detected metabolite peaks was performed by searching the NIST08 mass spectral library. The compounds were quantified from the peak area relative to the IS ribitol. No response factors were calculated. The representative GC-MS spectra for yeast metabolic profile are shown in Figure 4.
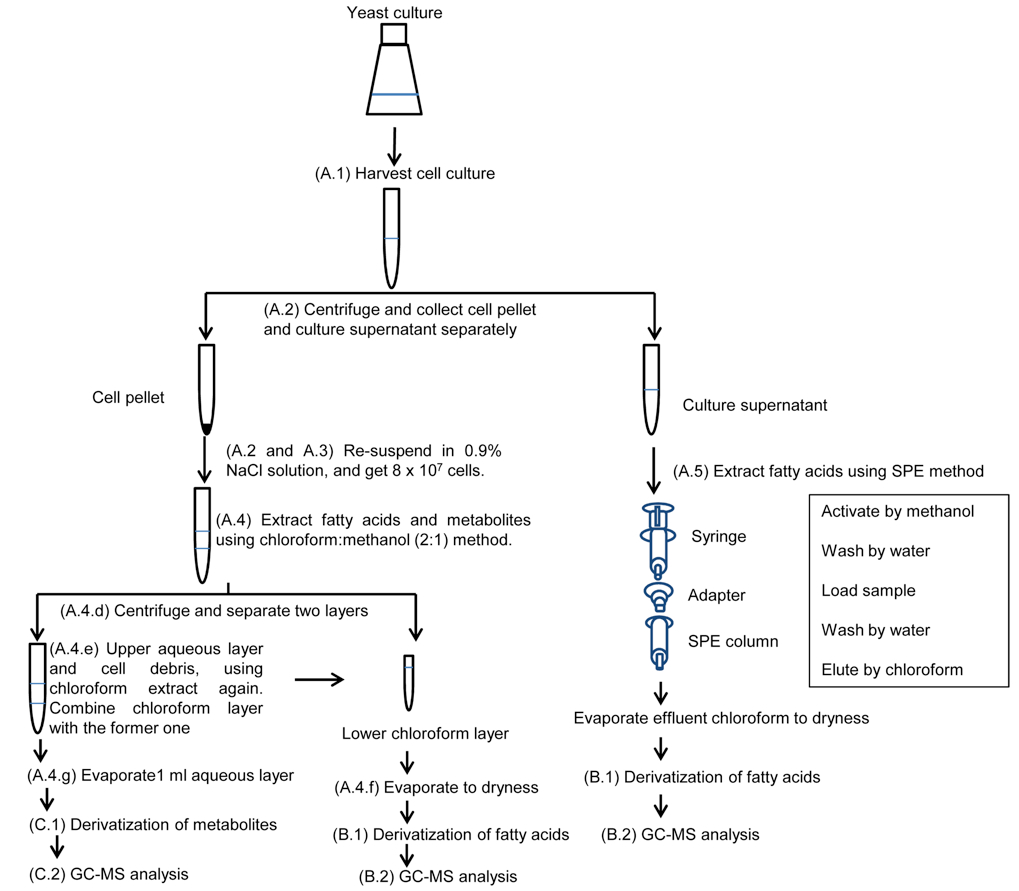
Figure 1. Workflow of metabolite and fatty acid analysis of yeast cells and culture supernatants
- Samples of 1 µl were injected into the column by splitless mode using an autoinjector. Helium was used as a carrier gas at 1.1 ml/min. The inlets and MS source temperatures were maintained at 250 and 230 °C, respectively. The oven temperature was maintained at 75 °C for 4 min and raised at 4 °C/min to a final temperature of 280 °C and held for 2 min. Mass spectra were recorded from 35 to 600 m/z.
- The sample for metabolic profiling was derivatized according to previous work (Wang et al., 2010).
Representative data
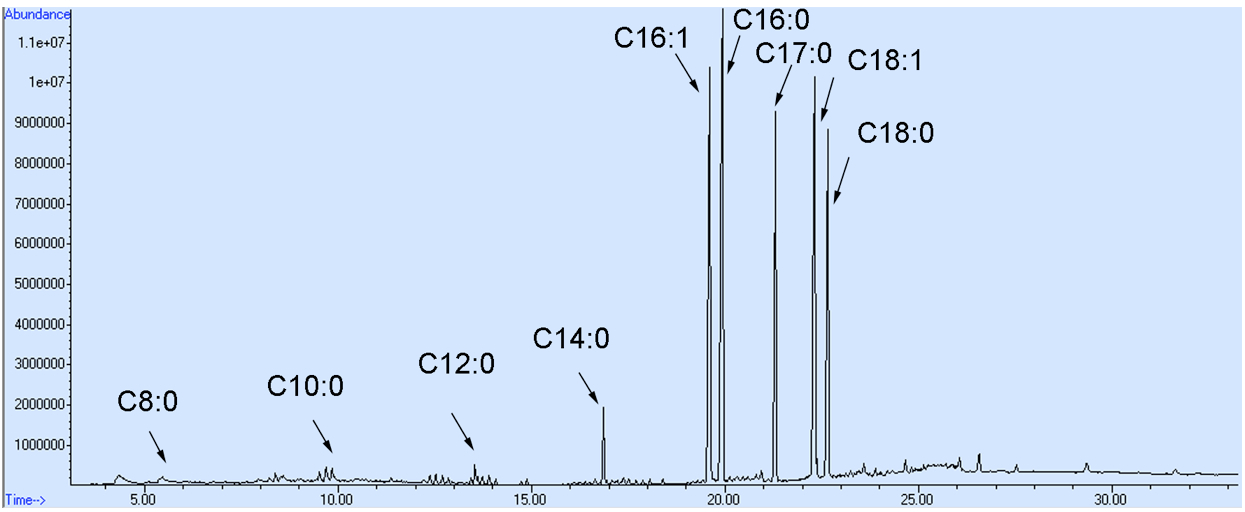
Figure 2. The representative GC-MS spectra for a S. cerevisiae fatty acid profile. Fatty acid composition and concentration may differ from those shown here as a result of altered cell culturing conditions and/or alternative genetic backgrounds of the yeast cells employed.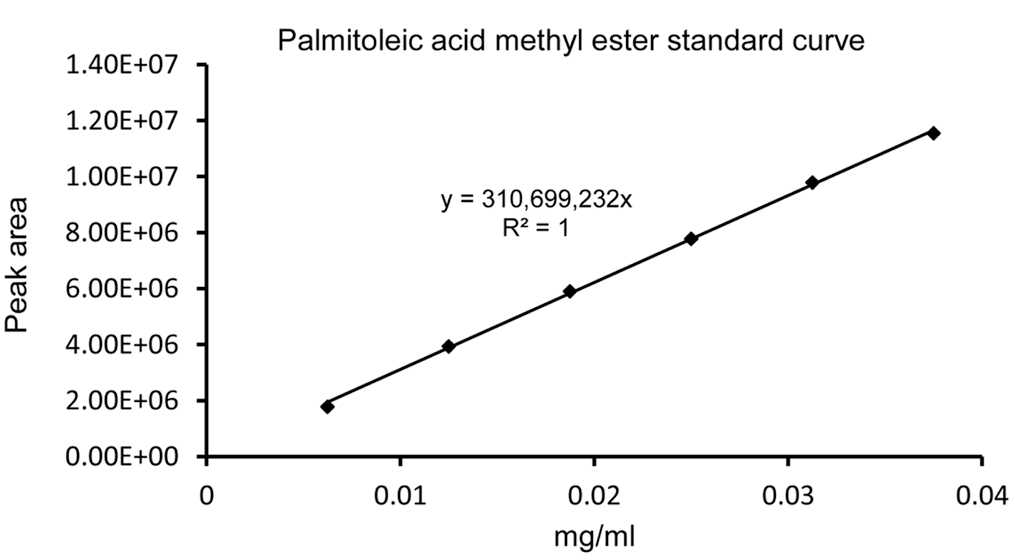
Figure 3. A representative palmitoleic acid methyl ester standard curve. Different concentrations (0.125-0.75 mg/ml) of FAMEs standard were used for GC-MS analysis. To make individual FAME standard curves, the concentration of FAME was determined by mutiplying its percent concentration (as indicated on the cerificate of analysis from the supplier) by the prepared concentrations (0.125-0.75 mg/ml).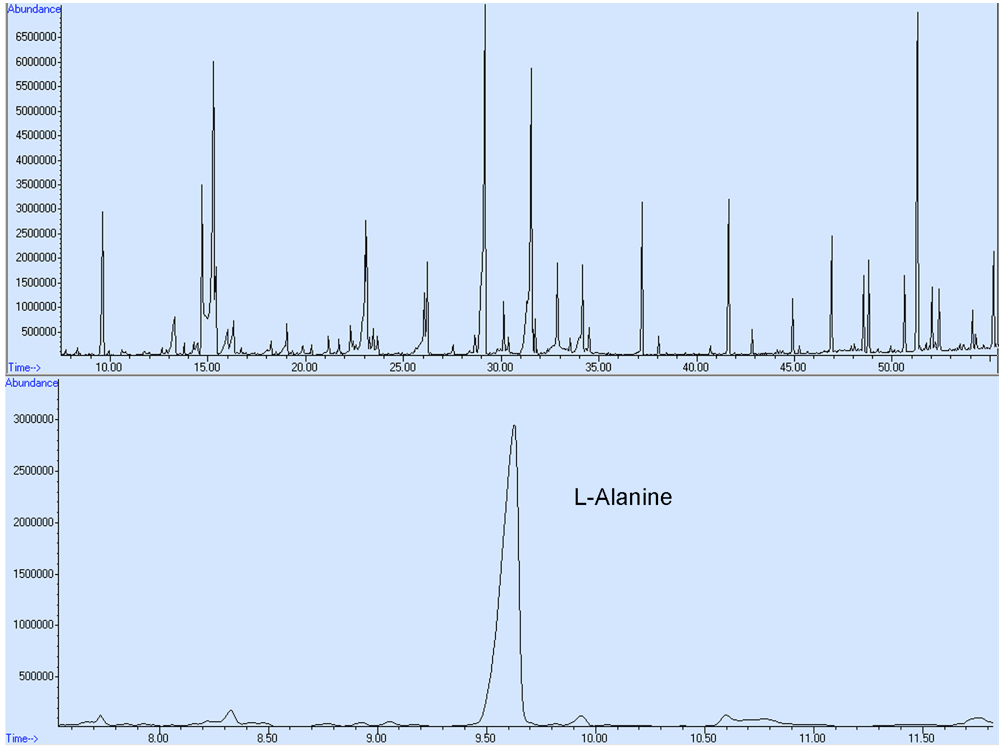
Figure 4. A representative GC-MS spectra for L-Alanine derived from total ion chromatograms. Intracellular metabolites were extracted from the S. cerevisiae cells and metabolic profiling was conducted. The metabolite composition and concentration may differ from those shown here as a result of altered cell culturing conditions and/or alternative genetic backgrounds of the yeast cells employed.
Notes
- For the metabolite and fatty acid analysis, at least three independent experiments were conducted.
- To ensure the reproducibility and comparability, please ensure the detector is well maintained and perform required maintenance for the machine, including cleaning the EI source and replacing the filament as necessary and in accordance with the manufacturers guidelines. For accurate comparisons, it is important that control and experimental cell samples be processed and analyzed in parallel.
- Adding or collecting organic solvent was carried out in the fume hood.
Recipes
- 10 mg/ml heptadecanoic acid (20 ml)
Mix 200 mg of heptadecanoic acid with 15 ml ethanol
Dissolved, add ddH2O to 20 ml
Stored at 4 °C - 10 mg/ml heptanoic acid
Mix 217.9 µl heptanoic acid with 15 ml ethanol
Dissolved, add ddH2O to 20 ml
Stored at 4 °C - 2 mg/ml ribitol (20 ml)
Mix 40 mg of ribitol with 15 ml ddH2O
Dissovled, add ddH2O to 20 ml
Stored at 4 °C - Saturated NaCl solution
Mix 35.7 g of NaCl with 80 ml of ddH2O
Dissolved, add ddH2O to 100 ml
Stored at room temperature
Acknowledgments
This protocol was adapted from previous work (Chen et al., 2014). This research is supported by a Competitive Research Programme (CRP) grant from the National Research Foundation of Singapore.
References
- Browse, J., McCourt, P. J. and Somerville, C. R. (1986). Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal Biochem 152(1): 141-145.
- Chen, L., Zhang, J. and Chen, W. N. (2014). Engineering the Saccharomyces cerevisiae β-oxidation pathway to increase medium chain fatty acid production as potential biofuel. PLoS One 9(1): e84853.
- Horak, T., Culik, J., Cejka, P., Jurkova, M., Kellner, V., Dvorak, J. and Haskova, D. (2009). Analysis of free fatty acids in beer: comparison of solid-phase extraction, solid-phase microextraction, and stir bar sorptive extraction. J Agric Food Chem 57(23): 11081-11085.
- Wang, M., Bai, J., Chen, W. N. and Ching, C. B. (2010). Metabolomic profiling of cellular responses to carvedilol enantiomers in vascular smooth muscle cells. PLoS One 5(11): e15441.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Chen, L. and Chen, W. N. (2014). Metabolite and Fatty Acid Analysis of Yeast Cells and Culture Supernatants. Bio-protocol 4(17): e1219. DOI: 10.21769/BioProtoc.1219.
Category
Microbiology > Microbial metabolism > Lipid
Cell Biology > Cell metabolism > Lipid
Biochemistry > Lipid > Lipid measurement
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










