- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Endolysin Expression, Purification and Activity Determination by Zymography
Published: Vol 4, Iss 16, Aug 20, 2014 DOI: 10.21769/BioProtoc.1208 Views: 15845
Reviewed by: Kanika GeraAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Monitoring Protein Stability In Vivo Using an Intein-Based Biosensor
John S. Smetana [...] Christopher W. Lennon
Apr 20, 2025 1575 Views

Endo-1,4-β-D-xylanase Assay Using Azo-Xylan and Variants Thereof
Luca Bombardi [...] Salvatore Fusco
Apr 20, 2025 1916 Views

Thermus thermophilus CRISPR Cas6 Heterologous Expression and Purification
Junwei Wei [...] Yingjun Li
Jul 20, 2025 2167 Views
Abstract
Endolysins are peptidoglycan-degrading (muralytic) enzymes produced by many bacteriophages for cell lysis of the host bacterium. The enzymatic activity of muralytic enzymes can be assayed qualitatively using a zymogram containing incorporated peptidoglycan. This protocol describes the expression of a recombinant 6x His-tagged endolysin using an Escherichia coli (E. coli) expression system and native affinity purification of the protein using Ni-NTA agarose. For the zymogram, the protocol details isolation of crude peptidoglycan from the Gram-negative bacterium Rhodobacter capsulatus and the zymography of purified protein and crude cell lysate. Construction of an E. coli BL21 (DE3) pET28-a(+)-derived endolysin-expression system is briefly described.
The protocol described here was developed and optimized for the endolysin 555 utilized by the Rhodobacter capsulatus bacteriophage-like gene transfer agent (RcGTA) (Westbye et al., 2013) and to study the muralytic activities of protein P14 of RcGTA (Fogg et al., 2012), but should be transferrable as a general protocol to express and study a variety of endolysins.
Materials and Reagents
- Protein purification
- pET28-a(+) (EMD Millipore, catalog number: 69864 ) or similar IPTG-inducible T7-based protein expression plasmid incorporating a 6x His tag
- Standard reagents and tools for molecular cloning (see Hasmann et al., 2011)
- E. coli BL21 (DE3) (New England BioLabs, catalog number: C2527I , or other source), or similar E. coli T7-based protein overexpression strain
- Kanamycin sulfate (50 mg/ml in dH2O, filter sterilized) or appropriate antibiotic if using different plasmid
- Isopropyl β-D-1-thiogalactopyranoside (IPTG) (100 mM in dH2O, filter sterilized)
- Ni-NTA Agarose (QIAGEN, catalog number: 30210 )
- Imidazole (2 M in dH2O)
- Lysogeny broth (LB) (see Recipes)
- Lysis buffer (see Recipes)
- pET28-a(+) (EMD Millipore, catalog number: 69864 ) or similar IPTG-inducible T7-based protein expression plasmid incorporating a 6x His tag
- SDS-PAGE and zymogram
- SDS-PAGE gel (12% separation and 4% stacking layer, He, 2011)
- Laemmli buffer/sample loading buffer (He, 2011)
- 2-propanol
- Renaturation buffer (see Recipes)
- Coomassie brilliant blue protein stain solution (see Recipes)
- YPS broth (see Recipes)
- Destain solution (see Recipes)
- Phosphate buffer (see Recipes)
- SDS-PAGE gel (12% separation and 4% stacking layer, He, 2011)
Equipment
- 250 ml culture flasks
- Incubator with shaker for culture flask (temperature adjustable) (or similar)
- French press for cell lysis (see Note on alternative lysis methods)
- Rotor JA-20 (Beckman Coulter) or equivalent
- Centrifuge suitable for rotor (Beckman Coulter, model: J2-HS or similar)
- Gravity column for Ni-NTA agarose
- SDS-PAGE apparatus with power supply (Mini-Protean and PowerPac, Bio-Rad Laboratories or equivalent)
- Microtube centrifuge (table-top)
- Microwave (optional)
- Flat (roux, or tissue culture) bottles (1 L for photoheterotrophic growth)
- Light-emitting incubator for photoheterotrophic growth (see Note 6)
- Erlenmeyer flask (500 ml) (pyrex, or other heat resistant glass)
- Glass beaker (1 L or large enough to encompass a 500 ml flask) (pyrex, or other heat resistant glass)
- Bunsen burner (or similar heat source)
- Vacuum concentrator (Savant SpeedVac SC110A concentrator with UVS400 vacuum system, or similar) (optional)
Procedure
- Construction of IPTG-inducible 6x His-tagged recombinant endolysin expression system
- The open reading frame of the putative endolysin is cloned into the vector pET28-a (+) using the unique NcoI (C-terminal 6x His tag) or NdeI (N-terminal 6x His tag) and XhoI restriction sites by standard molecular cloning methods. The resultant plasmid should encode a recombinant protein incorporating a terminal 6x His tag driven by a T7-based promoter (Note 1).
- Transform the resultant expression plasmid into E. coli BL21 (DE3) cells, to create an IPTG-inducible 6x His tagged endolysin expression system (Note 2).
- The open reading frame of the putative endolysin is cloned into the vector pET28-a (+) using the unique NcoI (C-terminal 6x His tag) or NdeI (N-terminal 6x His tag) and XhoI restriction sites by standard molecular cloning methods. The resultant plasmid should encode a recombinant protein incorporating a terminal 6x His tag driven by a T7-based promoter (Note 1).
- Native batch purification of 6x His tagged protein from E. coli
- Grow culture of BL21 (DE3) containing the expression plasmid in 10 ml LB containing appropriate antibiotic at 37 °C.
- Inoculate starter culture into 100 ml LB containing appropriate antibiotic in 250 ml culture flask to an optical density at 600 nm (OD600) of ~0.01 to 0.05. Incubate at 37 °C with shaking (200 rpm) to an OD600 of 0.5.
- Induce expression by addition of IPTG to 1 mM, shift culture to 30 °C (optional) and incubate for 4 h (200 rpm) (Notes 3 and 4).
- Harvest cells by centrifugation at 3,500 rcf at 4 °C and resuspend pellet in 2 to 5 ml lysis buffer.
- Lyse cells by passing through a French press at 900 psi, alternatively use sonicator. Clear lysate by centrifugation 16,000 rcf for 10 min and collect supernatant.
Note: A French press should only be operated by trained personnel. Ensure that maximum pressure for cell is not exceeded. - Add 1 to 3 ml Ni-NTA agarose to an empty gravity protein column. Wash with three column volumes of lysis buffer to equilibrate beads.
- Load cleared supernatant on column (see Figure 1), and allow to drain, collecting flow through. Wash with 5 column-volumes of lysis buffer.
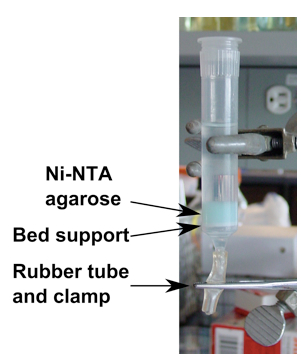
Figure 1. Column for Ni-NTA affinity purification. Gravity column for affinity purification using Ni-NTA agarose. Flow is controlled by clamping the short rubber tube on bottom of column. (The Ni-NTA agarose is stored in a solution of 20% ethanol.) - Elute protein with ten 0.5 to 1 ml aliquots lysis buffer supplemented with 450 mM imidazole (final concentration), collecting each fraction (Note 5).
- Run a standard SDS-PAGE gel containing crude lysate, cleared lysate, flowthrough, wash and eluates (typically 5 μl). Stain gel using Coomassie brilliant blue and destain using water or Destain solution.
Pool eluate fractions containing pure protein.
Note: This step is required to determine the purity of isolated protein. The eluate should contain a single band of predicted size. Presence of several bands requires optimization of binding or wash conditions, or could indicate proteolysis of the protein. Gels can be rapidly stained and destained by heating in microwave, taking care to avoid boiling.
- Grow culture of BL21 (DE3) containing the expression plasmid in 10 ml LB containing appropriate antibiotic at 37 °C.
- Isolation of crude peptidoglycan from R. capsulatus
- Inoculate R. capsulatus into 1 L capped (roux or tissue culture) flat bottles filled with YPS medium.
- Incubate photoheterotrophically at 30 °C with illumination until stationary phase (2 to 3 days) (Note 6). Harvest cells by centrifugation at 6,800 rcf and resuspend cell pellet in 100 ml 4% SDS in water.
- Boil suspension vigorously in heat-resistant glass flask by holding flask over Bunsen burner for 15 min, swirling occasionally. Keep beaker at hand.
Note: If suspension starts to foam and threaten to flow out of flask, move flask into beaker to collect and contain any spill. Wear appropriate safety equipment (goggles, gloves and lab coat advised as minimum). - Cool cell suspension, centrifuge at 10,000 rcf for 10 min. Gently decant supernatant.
- Wash pellet 5 times by resuspending in 10 ml dH2O followed by centrifugation (10,000 rcf 10 min).
- Resuspend pellet in a small volume dH2O (~3 ml).
Note: Crude peptidoglycan can be stored at 4 °C until use. - To estimate mass of purified peptidoglycan material, dry material in pre-weighted microcentrifuge tubes using a vacuum concentrator. Resuspend material in small volume dH2O.
- Inoculate R. capsulatus into 1 L capped (roux or tissue culture) flat bottles filled with YPS medium.
- Zymography of endolysin - protein separation
- Prepare SDS-PAGE zymogram by incorporating 5% (w/v) peptidoglycan into a standard 12% SDS-PAGE separating layer mixture before pouring into gel casting apparatus assembled according to manufacturer’s instructions. Overlay with 2-propanol and allow to set (20-40 min). Rinse 2-propanol off with dH2O, then overlay with standard 4% SDS-PAGE stacking layer and insert well-comb. Allow to set (20 - 40 min).
- Prepare (dilutions) of sample(s) by mixing with Laemmli loading buffer (5 μl Laemmli buffer per 10 μl sample).
- Briefly denature proteins by boiling for 5 min.
Note: It is advisable to perform an initial titration of denaturation temperature (and time) required for the protein to enter gel. Extensive heating (boiling) may prevent efficient protein re-folding required for the functional assay. - Load samples on gel and separate at 120 V until markers are sufficiently separated (~2 h). Dissemble gel apparatus, discard stacking layer.
- Prepare SDS-PAGE zymogram by incorporating 5% (w/v) peptidoglycan into a standard 12% SDS-PAGE separating layer mixture before pouring into gel casting apparatus assembled according to manufacturer’s instructions. Overlay with 2-propanol and allow to set (20-40 min). Rinse 2-propanol off with dH2O, then overlay with standard 4% SDS-PAGE stacking layer and insert well-comb. Allow to set (20 - 40 min).
- Zymography of endolysin - enzymatic assay
- Transfer resolving gel to plastic container, rinse extensively with dH2O to remove SDS-PAGE running buffer (Note 7).
- Renature proteins by washing gel twice in 5% Triton X-100 (v/v) in dH2O for 20 min with gentle shaking.
- For the enzymatic assay, decant Triton-X solution from gel and incubate the gel overnight (see Note 9) in 100 mM phosphate buffer at 30 °C or 37 °C with gentle shaking.
- To visualize, stain gels with Coomassie brilliant blue until bright blue. Destain using destain solution (Note 8).
- Enzyme activity is visible as a clearing against a blue background (see Figure 2).
Note: Clearing should only be observed at the correct molecular weight of the protein in samples of purified protein or crude lysate of cells expressing protein. Activity should be absent in crude lysate of empty vector-control strains.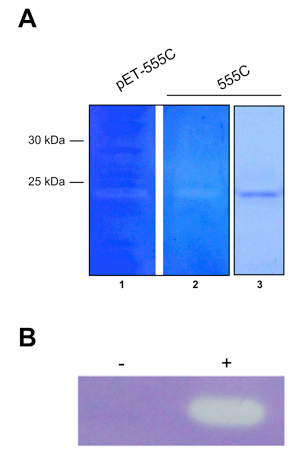
Figure 2. Zymograms of muralytic proteins. A. The endolysin 555 from R. capsulatus required for RcGTA release. Zymogram of E. coli total cell lysate (lane 1) and purified protein (lane 2), and SDS-PAGE gel of purified protein (lane 3). B. The muralytic enzyme P14 encoded in the RcGTA gene cluster. E. coli total cell lysate of empty vector control (-) and P14-expressing vector (+). Panel A is Copyright © American Society for Microbiology, Journal of Bacteriology, 195(22), 2013, p. 5025 - 5040 and DOI: 10.1128/JB.00669-13 and reused with permission from Reference 4. Panel B is adapted from Reference 1 in accordance with the Creative Commons Attribution License (CC BY).
- Transfer resolving gel to plastic container, rinse extensively with dH2O to remove SDS-PAGE running buffer (Note 7).
Notes
- Several other unique downstream restriction sites in addition to XhoI are present in pET28-a(+). For affinity purification using Ni-NTA, it is essential that the recombinant protein contains a 6xHis tag. The ATG-start codon of the endolysin gene should form part of the NdeI or NcoI restriction site. If using NcoI, the second codon may require optimization. The restriction sites are typically introduced in the PCR primer sequences. For DNA cloning work, we recommend the use of an E. coli strain optimized for cloning, such as DH5alpha.
- For high level T7-based expression of proteins such as the endolysin 555 from R. capsulatus in E. coli, the E. coli strain BL21 (DE3) or similar strains optimized for protein expression encoding IPTG-inducible T7 RNA polymerase works well. Be aware some E. coli protein expression strains contain the plasmids pLysS or pLysE, which express the T7 lysozyme (an endolysin).
- Expression conditions such as temperature should be determined empirically for each protein to be tested as many proteins are difficult to overexpress stably. For example, temperatures may be decreased to as low as 16 °C and IPTG induction concentrations can be decreased substantially.
- For initial expression, it is advisable to collect samples (1 ml) at several time points (0, 0.5, 1, 2 and 4 h) to optimize the time course of expression. Centrifuge and resuspend samples in Laemmli buffer and run on SDS-PAGE, followed by Coomassie brilliant blue staining to visualize.
- The binding affinity of proteins to Ni-NTA differs. The imidazole concentration required to elute a specific protein can be established by eluting protein using increasing concentration of imidazole. Run samples on SDS-PAGE and stain with Coomassie brilliant blue, to establish the concentration required to elute the protein from the Ni-NTA.
- A low cost incubator for photoheterotrophic growth of R. capsulatus can been constructed from a fishtank (“aquarium”) illuminated from the side by incandescent light bulbs, the temperature kept constant using a circulating water bath heater and immersing the culture vessel in the tank. Alternatively, R. capsulatus may be cultured chemoheterotrophically to lower cell density.
- A container lacking protruding internal edges is preferred to avoid damage to gel.
- If destained with water, the gel will destain slower, the color will change due to pH changes, and the intensity of background reduced.
- If no degradation is observed, possible reasons are:
- Absence of purified protein or cleavage of protein. Presence of purified protein of correct size should be verified by SDS-PAGE gel. If degradation/cleavage is expected, include protease inhibitors in lysis buffer.
- Too short incubation time. Increase incubation time.
- Low efficiency of protein renaturation. Subject samples to less intense heating before loading of gel, include low concentration (~0.5%) of Triton X-100 in phosphate buffer, or attempt using different detergent than Triton X-100 for renaturation. Alternatively, attempt a native gel using purified protein.
- Reducing agent may interfere with activity. Exclude beta-mercaptoethanol from Laemmli buffer.
- Absence of purified protein or cleavage of protein. Presence of purified protein of correct size should be verified by SDS-PAGE gel. If degradation/cleavage is expected, include protease inhibitors in lysis buffer.
Recipes
- Lysogeny broth (LB)
10 g Bacto tryptone
5 g yeast extract
10 g NaCl
Dissolved in 900 ml dH2O
Adjust pH to 7.0 using NaOH or HCl
Adjust to 1,000 ml final volume using dH2O
Autoclave - YPS broth
3 g yeast extract
3 g Bacto peptone
2 ml of 1 M MgSO4
2 ml of 1 M CaCl2
Dissolved in 900 ml dH2O
Adjust pH to 6.8 using NaOH or HCl
Adjust to 1,000 ml final volume using dH2O
Autoclave - Lysis buffer
20 mM NaCl
20 mM Tris-HCl (pH 8.0)
20 mM Imidazole - Renaturation buffer
5% (v/v) Triton X-100 in dH2O - Coomassie brilliant blue protein stain solution
0.1% (w/v) Coomassie brilliant blue R-250
10% (v/v) acetic acid (glacial)
40% (v/v) methanol
dH2O - Destain solution
10% (v/v) acetic acid (glacial)
40% (v/v) methanol
dH2O - Phosphate buffer (pH 7.5)
83.4 ml of 1 M K2HPO4
16.6 ml of 1 M KH2PO4
dH2O to 1,000 ml final volume
Note: Detailed SDS-PAGE gel buffers and sample loading/Laemmli buffers are described in He (2011).
Acknowledgments
The zymogram protocol was originally developed from Rosenthal and Dziarski (1994) and Hasmann et al. (2011).
References
- Fogg, P. C., Westbye, A. B. and Beatty, J. T. (2012). One for all or all for one: heterogeneous expression and host cell lysis are key to gene transfer agent activity in Rhodobacter capsulatus. PloS One 7(8): e43772.
- Hasmann, A., Wehrschuetz-Sigl, E., Kanzler, G., Gewessler, U., Hulla, E., Schneider, K. P., Binder, B., Schintler, M. and Guebitz, G. M. (2011). Novel peptidoglycan-based diagnostic devices for detection of wound infection. Diagn Microbiol Infect Dis 71(1): 12-23.
- He, F. L. (2011). Standard DNA Cloning. Bio-protocol 1(7): e52.
- He, F. L. (2011). Laemmli-SDS-PAGE. Bio-protocol 1(11): e80.
- Rosenthal, R. S. and Dziarski, R. (1994). Isolation of peptidoglycan and soluble peptidoglycan fragments. Methods Enzymol 235: 253–285.
- Westbye, A. B., Leung, M. M., Florizone, S. M., Taylor, T. A., Johnson, J. A., Fogg, P. C. and Beatty, J. T. (2013). Phosphate concentration and the putative sensor kinase protein CckA modulate cell lysis and release of the Rhodobacter capsulatus gene transfer agent. J Bacteriol 195(22): 5025-5040.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Westbye, A. B., Fogg, P. C. and Beatty, J. T. (2014). Endolysin Expression, Purification and Activity Determination by Zymography. Bio-protocol 4(16): e1208. DOI: 10.21769/BioProtoc.1208.
Category
Microbiology > Microbial biochemistry > Protein > Activity
Microbiology > Microbial biochemistry > Protein > Isolation and purification
Biochemistry > Protein > Expression
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











