- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Fluorometric Estimation of Viral Thermal Stability
Published: Vol 4, Iss 15, Aug 5, 2014 DOI: 10.21769/BioProtoc.1199 Views: 13202
Reviewed by: Kanika GeraAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Detection of HBV C Protein Phosphorylation in the Cell
Jaesung Jung and Kyongmin Kim
Aug 5, 2015 8580 Views

Host-regulated Hepatitis B Virus Capsid Assembly in a Mammalian Cell-free System
Kuancheng Liu and Jianming Hu
Apr 20, 2018 7097 Views

Detection and Analysis of S-Acylated Proteins via Acyl Resin–Assisted Capture (Acyl-RAC)
Dina A. Abdulrahman and Michael Veit
Apr 5, 2025 1837 Views
Abstract
Differential Scanning Fluorimetry (DSF) is a rapid, economical, and a straightforward technique for estimating the thermal stability of proteins. The principle involves the binding of a fluorescent dye to thermally exposed hydrophobic pockets of a protein. The dyes used in this technique are highly fluorescent in a non-polar environment and are quenched when exposed to aqueous solution. The change in fluorescence can be used to follow unfolding of proteins induced by temperature, pH, or chaotropic agents. The method is well characterized for monomeric proteins. Here, we extend the application to supramolecular protein and nucleo-protein complexes using virus particles as an example. SYPRO-orange™ dye is the dye of choice because it is matched for use with q-PCR instruments and the fluorescence response is stable across a wide range of pH and temperatures. Advantages of this technique over standard biophysical methods include the ability for high-throughput screening of biological and technical replicates and the high sensitivity.
Keywords: BiophysicsMaterials and Reagents
- Purified Virus Like Particles (VLPs) of Adeno-associated virus serotypes 1, 2, 5, 8
- Purified recombinant AAV (rAAV) capsids packaging a green fluorescent protein gene (GFP), rAAV1-GFP, rAAV5-GFP, rAAV8-GFP
- PBS (pH 7.0) at 25 °C
- Citric acid (Thermo Fisher Scientific, catalog number: BP339-500 )
- Disodium phosphate (VWR International, catalog number: BDH4538-1KGP )
- 5,000x SYPRO Orange (Life Technologies, catalog number: S6651 )
- Sodium hydroxide (Thermo Fisher Scientific, catalog number: BP359-500 )
- 12 N hydrochloric acid (EMD Millipore, catalog number: HX0603-3 )
- Sodium chloride (VWR International, catalog number: BDH4534-5KGP )
- Magnesium chloride (Thermo Fisher Scientific, catalog number : BP214-500 )
- Molecular biology grade water
- Citrate-Phosphate buffer (see Recipes)
Equipment
- Rotor Gene-3000, qPCR instrument with 36-well rotor (QIAGEN)
- 0.2 ml PCR tubes (BioExpress, catalog number: T-3013-1FC )
- Aluminum foil
- Pipette tips (10 µl, 200 µl) (VWR International, catalog numbers: 37001-162 and 53508-810 )
- Pipetts (200 µl, 0.1-2.5 µl) (Eppendorf, BioExpress, catalog numbers: P-3015-6 and P-3015-1 )
- pH meter (Accument Basic AB 15 pH meter) (Thermo Fisher Scientific)
- Milli-Q purification system (EMD Millipore)
Software
- Rotor gene software (Version 1.7, Build 94)
- Microsoft Excel 2007 on Windows 7
Procedure
- Each of the VLP and recombinant virus samples were diluted into PBS or the citrate-phosphate buffer to obtain concentrations between 0.1-0.24 mg/ml. A sample of lysozyme, at a concentration of 0.24 mg/ml, was included in each run as a positive control. The concentrations of the samples were varied to obtain similar fluorescence intensities on the instrument.
- Aliquots of the concentrated 5,000x SYPRO Orange was stored at -20 °C and gently and completely thawed out to room temperature before every reaction setup.
- 1 µl of the SYPRO Orange was diluted into 100 µl of distilled water to obtain a 1% 50x working stock. As SYPRO Orange is light sensitive, the working stock was covered with aluminum foil and kept in the dark till right before distribution.
- 2.5 µl of the working stock was added to each of the above samples. The remaining working stock, if any, was discarded if not used after 4 h.
- The samples were kept in an aluminum foil covered box to prevent exposure to light till they were transferred into the qPCR machine.
- The assays were conducted with a temperature ramping from 30 to 99 °C, increasing 1 degree every 1 min.
- The data was exported into a text (.csv) file and transferred into Microsoft Excel 2007 for data analysis.
Representative data
- The raw intensity data from all the samples was normalized to an upper limit of 100% by dividing all values by the largest recorded intensity in that sample. The data was plotted with temperature on the X-axis and the normalized intensity on the Y-axis.
- The raw data was also used to calculate the change in fluorescence per change in temperature (dF/dT). The maximum value obtained from this calculation was defined as the melting temperature (Tm). This can be accomplished by taking the difference between sequential fluorescent readings divided by the change in temperature between measurements. The change in intensity (dF) can then be graphed vs temperature. The largest value will can then be visualized or selected from the list. As described here, the temperature step-size is 1 degree.
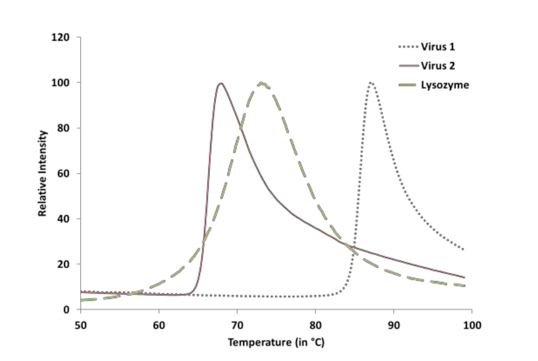
Figure 1. Showing a representative image of two Adeno-associated virus samples and a Lysozyme positive control. The x-axis shows temperature and y-axis shows the fluorescence intensity normalized to a 100%.
Notes
- SYPRO Orange can be excited with UV light at 300 nm or using visible light at 472 nm. The emission is measured 570 nm.
- The qPCR machine had a limited set of filters for SYPRO Orange. Though the excitation was possible at 470 nm, the spectra were collected at an emission wavelength of 550 nm. Although this was 20 nm lesser than the optimal emission wavelength, the peak intensity observed was ~50% of that observed at 570 nm. More importantly, the signal to noise ratio was very high thereby implying high quality data.
- The complementary data obtained from Differential Scanning Calorimetry (data not shown) also correlated accurately with that obtained from our method thereby further enhancing the value of our data (Rayaprolu et al., 2013).
- There are 36 vacant positions available on the rotor of the machine. A PCR tube that contains only buffer solution and the dye is used as a negative control. For this sample to be recognized as the negative control, this PCR tube needs to be placed in the first position in the rotor. The second position is occupied by a tube containing only buffer with no protein or dye added. This lets us determine if the buffer solution is contributing to the fluorescence intensity. The third position contains a lysozyme reaction aliquot (as described in the Procedure section). This acts as a positive control. Lysozyme’s melting temperature has been well documented in the literature. We compare the results we got from the DSF with the previously documented ones. More than one concentration of lysozyme was initially tested to confirm the melting temperature and the fluorescence linearity.
Recipes
- Citrate-Phosphate buffer
43.6% 0.2 M disodium phosphate
6.4% of 0.1 M citric acid
50 mM sodium chloride
Check to see if the pH is 7.0, if not, adjust with hydrochloric acid or sodium hydroxide to attain pH 7.0.
Make up to the required volume with water
Acknowledgments
The work was funded by the grant R01 AI081961-01A1 from National Institutes of Health (NIH). This protocol was adapted from Rayaprolu et al. (2012).
References
- Rayaprolu, V., Kruse, S., Kant, R., Venkatakrishnan, B., Movahed, N., Brooke, D., Lins, B., Bennett, A., Potter, T., McKenna, R., Agbandje-McKenna, M. and Bothner, B. (2013). Comparative analysis of adeno-associated virus capsid stability and dynamics. J Virol 87(24): 13150-13160.
- Vedadi, M., Niesen, F. H., Allali-Hassani, A., Fedorov, O. Y., Finerty, P. J., Jr., Wasney, G. A., Yeung, R., Arrowsmith, C., Ball, L. J., Berglund, H., Hui, R., Marsden, B. D., Nordlund, P., Sundstrom, M., Weigelt, J. and Edwards, A. M. (2006). Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proc Natl Acad Sci U S A 103(43): 15835-15840.
- Yeh, A. P., McMillan, A. and Stowell, M. H. (2006). Rapid and simple protein-stability screens: application to membrane proteins. Acta Crystallogr D Biol Crystallogr 62(Pt 4): 451-457.
- http://www.lifetechnologies.com/us/en/home/life-science/cell-analysis/labeling-chemistry/fluorescence- spectraviewer.html#product=S6650.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Rayaprolu, V., Kruse, S., Kant, R., Movahed, N., Brooke, D. and Bothner, B. (2014). Fluorometric Estimation of Viral Thermal Stability. Bio-protocol 4(15): e1199. DOI: 10.21769/BioProtoc.1199.
Category
Microbiology > Microbial biochemistry > Protein > Modification
Biochemistry > Protein > Modification
Biochemistry > Protein > Structure
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










