- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantification of Flavin Production by Bacteria
Published: Vol 4, Iss 15, Aug 5, 2014 DOI: 10.21769/BioProtoc.1197 Views: 11003
Reviewed by: Tie LiuFeng LiZhaohui LiuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Quantification of Colibactin-associated Genotoxicity in HeLa Cells by In Cell Western (ICW) Using γ-H2AX as a Marker
Sophie Tronnet and Eric Oswald
Mar 20, 2018 8074 Views

Adhesion of Enteroaggregative E. coli Strains to HEK293 Cells
Jorge Luis Ayala-Lujan and Fernando Ruiz-Perez
Apr 20, 2018 6850 Views

Intracellular Invasion and Killing Assay to Investigate the Effects of Binge Alcohol Toxicity in Murine Alveolar Macrophages
Victor Jimenez Jr and Fernando P Monroy
Jan 20, 2019 6735 Views
Abstract
This protocol provides a simple and fast method of quantification for intracellular flavin content, and for flavin secretion by bacteria. Intracellular flavins are extracted from bacterial pellets, and secreted flavins are examined in the cell growth medium. Flavins are separated and measured using HPLC with fluorescence detection, and quantified based on a comparison to standards.
Keywords: Riboflavin Biosynthetic PathwayMaterials and Reagents
- Sinorhizobium meliloti (S. meliloti 1021, Galibert et al., 2001)
Note: The protocol can be also applied to other bacteria. - Bio-Rad Protein Assay Kit I (Sigma-Aldrich, catalog number: 500-0001 )
- Flavin standards:
- Riboflavin (Sigma-Aldrich, catalog number: 95170 )
- FMN (Sigma-Aldrich, catalog number: F2253 )
- FAD (Sigma-Aldrich, catalog number: F6625 )
- Lumichrome (Acros, catalog number: 146930010 )
- Riboflavin (Sigma-Aldrich, catalog number: 95170 )
- Ammonium formate
- Formic acid
- Methanol
- YMB medium (Somerville and Kahn, 1983) (see Recipes) (1 plate per sample)
- MMNH4 medium (Somerville and Kahn, 1983) (see Recipes) (6 ml per sample)
- Extraction buffer (see Recipes)
- HPLC mobile phase (see Recipes)
Note: Except as otherwise noted, all other chemicals were obtained from Sigma-Aldrich.
Equipment
- Aluminum foil
- Eppendorf 1.7 ml tubes (Thermo Fisher Scientific, catalog number: 14-222-168 )
- 14 ml Falcon tubes (BD Biosciences, catalog number: 352059 )
- 0.22 µm syringe filter for HPLC sample preparation (Microsolv Technology, catalog number: 58022-N04-C )
- Scale (Ohaus Corporation, model: E10640 )
- Mini-centrifuge
- Shaker
- HPLC: Waters Alliance 2695 HPLC system linked to a 2475 fluorescence detector
- SunFire C18 reverse-phase column (4.6 x 150 mm, 3.5 µm)
Procedure
- Cell growth
- Grow S. meliloti on YMB plate for 48 h at 30 °C. The optimal temperature for S. meliloti growth is between 28 °C and 30 °C.
- Inoculate S. meliloti from the stock YMB plate to an OD600 of ~0.1 in 3 ml MMNH4 medium.
- Grow cells for 48 h at 30 °C, 250 rpm in 14 ml Falcon or glass tubes completely wrapped with foil to prevent light-induced flavin degradation. In case of testing flavin levels in light-grown culture or flavin degradation by light, the foil could be omitted.
- Dilute the cells 20-fold into 3 ml fresh MMNH4 medium and grow for 1 or 3 days at 30 °C, 250 rpm in 14 ml Falcon or glass tubes covered with aluminum foil. The length of cell growth could vary from several hours to 10+ days depending on the specific question of the research.
- Remove 1 ml of the growing culture for protein quantification. These 1 ml samples can be frozen at -20 °C for later protein quantification. (The rest 2 ml of cell cultures will be used for flavin detection, steps B, C and D.)
- For protein assay, break the cells by sonication (Fisher Sonic Dismembrator 300 with the intermediate size attachment and a power setting of 60 %, three times for 3 min each at 4 °C).
- Remove cells debris by centrifugation (≥ 3,000 x g; room temperature; 20 min).
- Measure protein concentration in the cultures by using Bio-Rad Protein Assay Kit.
Note: Carry out the rest of the procedure for flavin extraction and analysis under as limited lighting as possible since flavins are extremely light sensitive. Protect extracted and filtered flavins from light by storing in a light-tight container. For long-term storage, samples can be stored below 0 °C in a light-tight container.
- Grow S. meliloti on YMB plate for 48 h at 30 °C. The optimal temperature for S. meliloti growth is between 28 °C and 30 °C.
- Separation of cells from conditioned media
- Harvest cells from 1 ml culture by centrifugation (≥ 10,000 x g; room temperature; 20 min) in pre-weighed Eppendorf tubes.
- Transfer the supernatant to clean Eppendorf tubes. It will be used in step C and can be stored at -20 °C in foil wrapped Eppendorf tubes for later analysis.
- Wash the pellets with 1 ml MMNH4 buffer and centrifuge (≥ 10,000 x g; room temperature; 20 min).
- Pipet out all liquid and re-weigh the tube to measure the pellet mass. The pellet mass is expected to be at the range between 5 mg and 10 mg depending on the time of growth.
- The pellet will be used in step D can be stored at -20 °C in foil wrapped Eppendorf tubes for later analysis.
- Harvest cells from 1 ml culture by centrifugation (≥ 10,000 x g; room temperature; 20 min) in pre-weighed Eppendorf tubes.
- To measure secreted flavins
- Filter the supernatant left after harvesting the bacteria (step B) using Microsolv 0.22 µm syringe filters.
- For riboflavin, FMN, and FAD detection, separate the samples by reverse-phase chromatography using a Waters Alliance 2695 HPLC system with a Waters SunFire C18 column (4.6 x 150 mm, 3.5 µm) maintained at 35 °C linked to a Waters 2475 fluorescence detector. Detect riboflavin, FMN, and FAD by using an excitation wavelength of 470 nm and an emission wavelength of 530 nm. Run mobile phase at a flow rate of 1 ml/min for 12 min per sample. Maintain the samples at 10 °C.
- For lumichrome detection, use excitation and emission wavelengths of 260 nm and 470 nm respectively. Use the mobile phase gradient program indicated in Table 1. The column and flow rate are the same as indicated above. Run mobile phase for 15 min. Column temperature 35 °C.
- Determine the flavin concentration by comparison to standards (0.05, 0.1, 0.5, 1.0 and 5.0 µM FAD, FMN and riboflavin).
- Normalize the flavin concentration against the protein concentration (as calculated above in step A8).
Table 1. Solvent gradient program parameters for lumichrome detection*Flow rate 1 ml/minTime (min) Water (%) Methanol (%) 0 77 23 8 40 60 11 40 60 12 0 100 13 0 100 14 77 23 15 77 23
- Filter the supernatant left after harvesting the bacteria (step B) using Microsolv 0.22 µm syringe filters.
- To measure intracellular flavin concentration
- Resuspend the cell pellets from step B in extraction buffer (1: 10 w/v), heat at 80 °C for 10 min, then centrifuge at 20,000 x g for 20 min at 4 °C.
- Filter the supernatants using a 0.22 µm syringe filter.
- Measure riboflavin, FMN, FAD, and lumichrome as described for the secreted flavins.
- Resuspend the cell pellets from step B in extraction buffer (1: 10 w/v), heat at 80 °C for 10 min, then centrifuge at 20,000 x g for 20 min at 4 °C.
Representative data
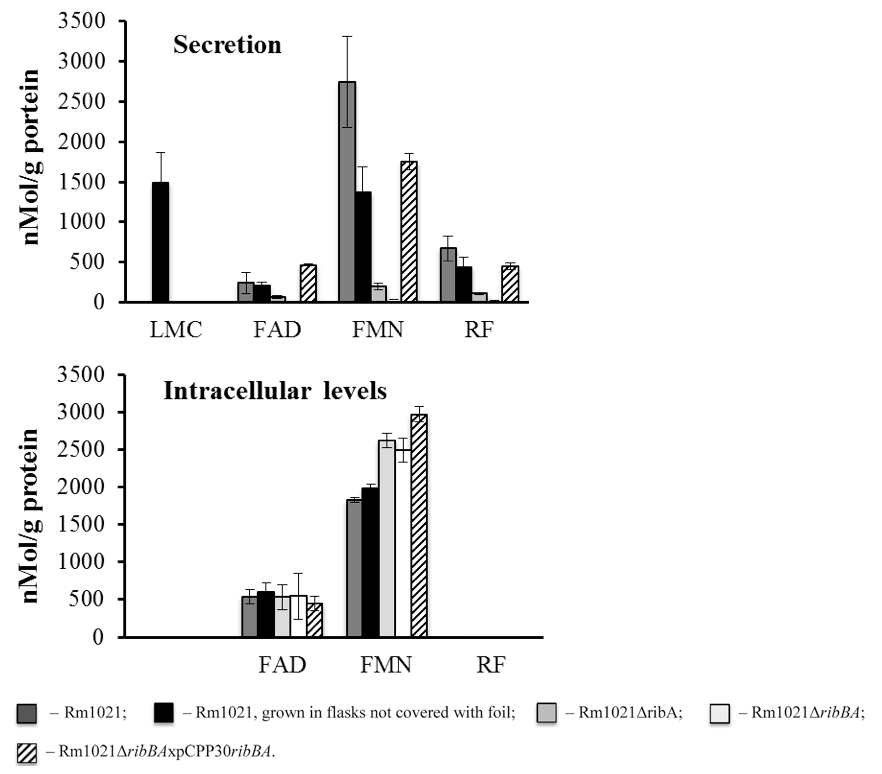
Figure 1. Flavin secretion and accumulation by the S. meliloti strains. S. meliloti strains were grown in MMNH4 media. 1 ml of cell cultures was taken after 3 days of growth. The cells were spun down and the mass of the pellets was measured. Lumichrome (LMC), FMN, FAD and riboflavin (RF) concentration in the supernatants and the pellets were measured using HPLC. The flavin concentration was normalized against the protein concentration. Rm1021 - wild type strain; Rm1021ΔribA and Rm1021ΔribBA - Rm1021 mutants with decreased ability to secret flavins; Rm1021ΔribBAxpCPP30ribBA - Rm1021 mutant with restored ability to secret flavins. Data are averaged from at least three independent experiments.
Recipes
- YMB media for Rhizobium
1 L Concentration Yeast Extract 1 g Mannitol 10 g 54.9 mM Agar 15 g
Autoclave, cool to 55 °C, then addYMB Salt I 10 ml YMB Salt II 10 ml YMB Salt I 1 L Concentration K2HPO4 50 g 287.06 mM NaCl 10 g 171.15 mM d-H2O 960 ml YMB Salt II 1 L Concentration MgSO4.7H2O 20 g 81.11 mM d-H2O 1 L - MMNH4 (minimal mannitol ammonia media for Rhizobium)
per 1 L Concentration Mannitol 10.0 g 54.9 mM NH4Cl 0.5 g 9.34 mM Agar (for plates preparation) 15.0 g d-H2O 970 ml
Autoclave, cool to 55 °C, then add:Biotin (0.2 mg/ml in 50% EtOH) 1.0 ml Thiamine (2 mg/ml), filter sterilized 1.0 ml Min Man Salts I 10.0 ml Min Man Salts II 10.0 ml
Min Man Salts Iper 1 L Concentration K2HPO4 100 g 574.12 mM KH2PO4 100 g 734.8 mM Na2SO4 25 g 174.8 mM d-H2O 1 L
Min Man Salt IIper 1 L Concentration FeCl3.6H2O 1.0 g 3.7 mM Concentrated HCl adjust pH to ~7.0 (~1 drop) CaCl2.2H2O 10.0 g 68 mM MgCl2.6H2O 25.0 g 123 mM d-H2O 1 L Autoclave - Extraction buffer
100 mM ammonium formate
100 mM formic acid
25% methanol - HPLC mobile phase
100 mM ammonium formate
100 mM formic acid
25% methanol
For 1 liter of extraction buffer/ mobile phase, add the following components in the order below. Mix well after adding all the components.550 ml water 100 ml ammonium formate, 1 M, filtered through a 0.22 μm filter 100 ml formic acid, ≥98% pure, 1 M 250 ml methanol, HPLC grade
Acknowledgments
This protocol was adapted from Yurgel et al., 2014. This work was supported by the Agricultural Research Center (WNP-00773) at Washington State University and grant DE-FG0396ER20225 Vol. 27, No. 5, 2014 / 445 from the Energy Biosciences Program at the United States Department of Energy, and by grant NSF-MCB 1052492 to S. Rajamani. This activity was funded, in part, with an Emerging Research Issues Internal Competitive grant from the Agricultural Research Center at Washington State University, College of Agricultural, Human, and Natural Resource Sciences and Biologically-Intensive Agriculture and Organic Farming (BIOAg) Internal Competitive grant from The Center for Sustaining Agriculture and Natural Resources (CSANR) at Washington State University to S. Yurgel. We thank M. Kahn for discussions and the Washington State University Laboratory for Biotechnology and Bioanalysis for sequencing support.
References
- Galibert, F., Finan, T. M., Long, S. R., Puhler, A., Abola, P., Ampe, F., Barloy-Hubler, F., Barnett, M. J., Becker, A., Boistard, P., Bothe, G., Boutry, M., Bowser, L., Buhrmester, J., Cadieu, E., Capela, D., Chain, P., Cowie, A., Davis, R. W., Dreano, S., Federspiel, N. A., Fisher, R. F., Gloux, S., Godrie, T., Goffeau, A., Golding, B., Gouzy, J., Gurjal, M., Hernandez-Lucas, I., Hong, A., Huizar, L., Hyman, R. W., Jones, T., Kahn, D., Kahn, M. L., Kalman, S., Keating, D. H., Kiss, E., Komp, C., Lelaure, V., Masuy, D., Palm, C., Peck, M. C., Pohl, T. M., Portetelle, D., Purnelle, B., Ramsperger, U., Surzycki, R., Thebault, P., Vandenbol, M., Vorholter, F. J., Weidner, S., Wells, D. H., Wong, K., Yeh, K. C. and Batut, J. (2001). The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293(5530): 668-672.
- Sandoval, F. J., Zhang, Y. and Roje, S. (2008). Flavin nucleotide metabolism in plants: monofunctional enzymes synthesize fad in plastids. J Biol Chem 283(45): 30890-30900.
- Somerville, J. E. and Kahn, M. L. (1983). Cloning of the glutamine synthetase I gene from Rhizobium meliloti. J Bacteriol 156(1): 168-176.
- Yurgel, S. N., Rice, J., Domreis, E., Lynch, J., Sa, N., Qamar, Z., Rajamani, S., Gao, M., Roje, S. and Bauer, W. D. (2014). Sinorhizobium meliloti flavin secretion and bacteria-host interaction: role of the bifunctional RibBA protein. Mol Plant Microbe Interact 27(5): 437-445.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Yurgel, S. N., Lynch, J., Rice, J., Adhikari, N. and Roje, S. (2014). Quantification of Flavin Production by Bacteria. Bio-protocol 4(15): e1197. DOI: 10.21769/BioProtoc.1197.
Category
Microbiology > Microbe-host interactions > In vitro model > Cell line
Microbiology > Microbial biochemistry > Other compound
Plant Science > Plant physiology > Endosymbiosis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









