- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Murine in vivo CD8+ T Cell Killing Assay
Published: Vol 4, Iss 13, Jul 5, 2014 DOI: 10.21769/BioProtoc.1172 Views: 19628
Reviewed by: Omar AkilAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Visualizing Hypoxia in a Murine Model of Candida albicans Infection Using in vivo Biofluorencence
José Pedro Lopes and Constantin F. Urban
Aug 5, 2019 5104 Views

RNA Extraction from Ears and Draining Lymph Nodes of Mice Infected with Leishmania amazonensis
Emilie Giraud and Evie Melanitou
Jun 5, 2020 5513 Views

TetR Regulated in vivo Repression Technology to Identify Conditional Gene Silencing in Genetically Engineerable Bacteria Using Vibrio cholerae Murine Infections as Model System
Franz G. Zingl [...] Stefan Schild
Oct 5, 2020 3743 Views
Abstract
Antigen-specific killing ability of effector CD8+ T cells is critical for protective immunity against infection. Here, we describe in vivo cytotoxic T cell assay to examine effector function of antigen-specific CD8+ T cells. Mice infected with Listeria monocytogenes (L. monocytogenes) expressing chicken ovalbumin as a model antigen mount ovalbumin-specific CD8+ T cell responses. Effector CD8+ T cell function in vivo is determined by mixed transfer of OVA peptide-pulsed target cells with control target cells into the previously immunized mice. Difference in CFSE expression levels clearly marks two distinct populations: Antigen-pulsed target cells-CFSElow vs. unpulsed target cells-CFSEhi. The frequencies between antigen-pulsed target cells and control target cells are used as readouts of antigen-specific killing.
Materials and Reagents
- Splenocytes from a wild type mouse
- PBS (Thermo Fisher Scientific, catalog number: BP399-20 )
Note: 10x solution, diluted to 1x in house in distilled water and sterilized by autoclave.
- RBC lysis buffer (eBioscience, catalog number: 00-4333-57 )
- HBSS without Ca2+ and Mg2+ (Life Technologies, Gibco®, catalog number: 14175-095 )
- RPMI-1640 medium (Life Technologies, Gibco®, catalog number: 11875-119 )
- Fetal bovine Serum (Atlanta Biologicals, catalog number: S11055H )
- Penicillin/streptomycin (Gemini Bio-Products, catalog number: F52M00E )
- L-Glutamine (Life Technologies, Gibco®, catalog number: 25030-081 )
- Trypan blue solution (Life Technologies, Gibco®, catalog number: 15250-061 )
- OVA257-264 synthetic peptide (Sigma-Aldrich, catalog number: S7951 )
- 5(6)-Carboxyfluorescein diacetate N-succinimidyl ester (CFSE) (Sigma-Aldrich, catalog number: 21888 )
- Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D-8418 )
- Collagenase D (Sigma-Aldrich, catalog number: C-5138 )
- Percoll (Sigma-Aldrich, catalog number: P-1644 )
- Complete RPMI-1640 media (see Recipes)
- 100% percoll solution (see Recipes)
Equipment
- Centrifuge (Thermo Fischer Scientific, SorvallTM Legend RT )
- 37 °C water bath
- Hemocytometer
- 15 ml and 50 ml Falcon tubes
- 6 well plates (USA Scientific, CytoOne®, catalog number: CC7682-7506 )
- BD LSRII Flow Cytometer (BD)
- 70 µm cell strainer (BD Biosciences, Falcon®, catalog number: 352350 )
- 5 ml polystyrene round-bottom tubes with cell-strainer cap (BD Biosciences, Falcon®, catalog number: 352235 )
- 3 ml syringe (BD, catalog number: 14-823-435 )
Procedure
- Target cell preparation under sterile tissue culture conditions
This step is for preparing peptide-pulsed target cells and stain cells with CFSE to distinguish peptide-pulsed target cells from control target cells.- OVA257-264 peptide-loading for the target cells.
- Splenocytes are RBC lysed followed by washing with PBS twice.
- Resuspend cells in RPMI-1640 complete medium.
- Count the mononuclear cells by trypan blue exclusion using a hemocytometer.
- Resuspend cells at 5 x 106/ml of RPMI-1640 complete medium.
- Divide the cells equally into two separate 50 ml Falcon tubes- one for peptide-pulsed target cells, and the other for unpulsed target cells.
- Add OVA257-264 peptide at 1 µl/ml from a 200 µM stock to peptide-pulsed target cells.
- Add an equivalent amount of PBS to the unpulsed target cells.
- Incubate the cells in a 37 °C water bath for 1 h.
- Wash cells twice with RPMI-1640 complete medium.
- Centrifuge the cells at 1,500 rpm for 3 min at 4 °C.
- Resuspend the cell pellet in HBSS.
- Splenocytes are RBC lysed followed by washing with PBS twice.
- CFSE cell labeling under sterile tissue culture conditions.
- Count all live cells by trypan blue exclusion using hemocytometer.
- Resuspend the cells in HBSS at 5 x 107/ml.
- Thaw an aliquot of 5 mM stock CFSE solution.
- Make a fresh CFSElow stock solution by diluting 5 mM stock 1:10 in DMSO (a final concentration of 0.5 mM).
- Incubate the unpulsed target splenocytes with the higher concentration of CFSE (CFSEhigh): Add 1 µl of the 5 mM stock CFSE for each milliliter of unpulsed target cells (final concentration of 5 µM).
- Incubate the pulsed target splenocytes with the lower concentration of CFSE (CFSElow): Add 1 µl of the 0.5 mM stock CFSE for each milliliter of peptide-pulsed cells (final concentration of 0.5 µM).
- Pipette cells up and down to mix well and incubate in water bath for 10 min at 37 °C. Gently agitate the cells periodically.
- Add 10x the volume of pre-warmed RPMI-1640 complete medium to the CFSE-labeled cells to stop the reaction.
- Pellet cells at 1,500 rpm for 3 min at 4 °C.
- Remove the supernatant and resuspend the pellet in cold RPMI-1640 complete medium.
- Wash the cells two more times with cold RPMI-1640 complete medium.
- Count the mononuclear cells by trypan blue exclusion using a hemacytometer.
- Wash the cells with cold PBS.
- Resuspend each cell populations in PBS at 6.7 x 106/ml.
- Combine an equal volume (~equal numbers) of peptide-pulsed CFSElow cells with unpulsed CFSEhi cells and proceed with flow cytometry analysis (Figure 1a).
- Count all live cells by trypan blue exclusion using hemocytometer.
- OVA257-264 peptide-loading for the target cells.
- Intravenous injection of target splenocytes
To investigate OVA257-264-specific CD8+ T cell killing ability, peptide-pulsed and unpulsed target cells were mixed at a 1:1 ratio and transferred to the previously immunized mice.- Recipient mice were infected with 5,000 colony-forming units (CFU) of Listeria monocytogenes expressing chicken ovalubmin (LM-OVA) 7 days intravenously before the CFSE-labeled cell injection.
- Inject intravenously 300 µl of the combined cell populations into the tail vein of each recipient. Each recipient should receive approximately 1 x 107 peptide-pulsed target cells combined with 1 x 107 unpulsed target cells.
- Wait for 4 h.
- Recipient mice were infected with 5,000 colony-forming units (CFU) of Listeria monocytogenes expressing chicken ovalubmin (LM-OVA) 7 days intravenously before the CFSE-labeled cell injection.
- Preparation of splenocytes and lymphocytes in the liver for flow cytometry analysis
This step is analyzing antigen-specific killing ability in the liver and the spleen by flow cytometric analysis of CFSElow and CFSEhi cell populations.- Prepare splenocytes for flow cytometry analysis.
- Wash once with PBS, and 2-300 µl into a 5 ml round bottom tubes through the cell strainer.
- Isolate lymphocytes from the liver.
- Harvest livers from the recipients and place them on ice.
- Make a fresh collagenase D solution by diluting 20 mg/ml stock 1:20 in PBS (a final concentration of 1 mg/ml).
- Chop liver with a blade on a slide glass and transfer them into a 50 ml falcon tube.
- Add 7 ml of collagenase D (1 mg/ml) and vortex well.
- Incubate in water bath for 30 min at 37 °C. Vortex every 15 min.
- Put tubes on ice and add supernatant on 70 µm filter on a well of a 6 well plate. Grind chunks of the chopped liver with flat portion of 3 ml syringe. Wash the tube with 5 ml of PBS and repeat grinding.
- Transfer them to a 50 ml Falcon tube and spin down at 2,000 rpm for 5 min at 4 °C.
- Make fresh 44% and 66% percoll solution: Make 44% final concentration by diluting 100% percoll in PBS, and make 66% final concentration by diluting 100% percoll in RPMI-1640 medium.
- Resuspend pellet in 7 ml of 44% percoll, and load them on 3 ml of 66% percoll in 15 ml tube.
Note: The gradient separation is sensitive to agitation. Try not to shake the tube.
- Centrifuge at 3,000 rpm for 30 min at 4 °C without brake.
- Transfer the interphase lymphocytes to a new 15 ml tube.
- Pellet cells at 2,000 rpm for 5 min at 4 °C.
- Wash once with PBS, and transfer 2-300 µl into a 5 ml round bottom tubes through the cell strainer.
- Harvest livers from the recipients and place them on ice.
- Proceed to flow cytometry analysis with spleen and liver samples (Figure 1b).
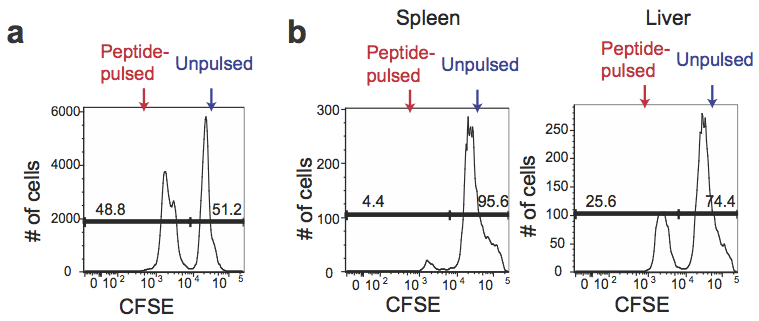
Figure 1. CFSE expression in antigen-pulsed target cells and unpulsed target cells. a. Peptide-pulsed CFSElow and unpulsed CFSEhi splenocytes were mixed at a 1:1 ratio before transferring to the recipients. b. The mixture of peptide-pulsed and unpulsed splenocytes was transferred into the mice predisposed with LM-OVA at day 7 post infection. The spleen and the liver of the recipients were harvested after 4 h to determine percentages of CFSElow and CFSEhi cells among CFSE+ cells.
- Prepare splenocytes for flow cytometry analysis.
Recipes
- Complete RPMI-1640 medium
10% FBS
1% Penicillin/streptomycin with L-Glutamine
- 100% percoll solution
90% of percoll
10% of 10x PBS
Notes
- The gradient separation is sensitive to agitation. Try not to shake the tube.
Acknowledgments
The protocol was adapted from a previously described study (Manjunath et al., 2001). This work was supported by the Starr Cancer Consortium (13-A123 to M.O.L. and M.Q.Z.), the Rita Allen Foundation (M.O.L.), the NBRPC (2012CB316503 to M.Q.Z), and the NIH (HG001696 to M.Q.Z.).
References
- Ingulli, E. (2007). Tracing tolerance and immunity in vivo by CFSE-labeling of administered cells. Methods Mol Biol 380: 365-376.
- Kim, M. V., Ouyang, W., Liao, W., Zhang, M. Q. and Li, M. O. (2013). The transcription factor Foxo1 controls central-memory CD8+ T cell responses to infection. Immunity 39(2): 286-297.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Kim, M. V., Ouyang, W., Liao, W., Zhang, M. Q. and Li, M. O. (2014). Murine in vivo CD8+ T Cell Killing Assay. Bio-protocol 4(13): e1172. DOI: 10.21769/BioProtoc.1172.
Category
Immunology > Immune cell function > Antigen-specific response > Membrane protein detection
Immunology > Animal model > Mouse
Microbiology > Microbe-host interactions > In vivo model > Mammal
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link