- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Production and Crystallization of Nanobodies in Complex with the Receptor Binding Domain of the SARS-CoV-2 Spike Protein
(*contributed equally to this work) Published: Vol 12, Iss 9, May 5, 2022 DOI: 10.21769/BioProtoc.4406 Views: 3703
Reviewed by: Joana Alexandra Costa ReisLuke A YatesSneha Ray

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Carbamoyltransferase Enzyme Assay: In vitro Modification of 5-hydroxymethylcytosine (5hmC) to 5-carbamoyloxymethylcytosine (5cmC)
Weiwei Yang [...] Laurence Ettwiller
Sep 5, 2022 1985 Views

Cell-Sonar, an Easy and Low-cost Method to Track a Target Protein by Expression Changes of Specific Protein Markers
Sabrina Brockmöller [...] Simone Rothmiller
Feb 5, 2025 1657 Views

Cluster FLISA—A Method to Compare Protein Expression Efficiency Between Cell Lines and Subunit Clustering of Proteins
Sabrina Brockmöller and Lara Maria Molitor
Nov 5, 2025 1207 Views
Abstract
The receptor binding domain (RBD) of the spike protein of SARS-CoV-2 binds angiotensin converting enzyme-2 (ACE-2) on the surface of epithelial cells, leading to fusion, and entry of the virus into the cell. This interaction can be blocked by the binding of llama-derived nanobodies (VHHs) to the RBD, leading to virus neutralisation. Structural analysis of VHH-RBD complexes by X-ray crystallography enables VHH epitopes to be precisely mapped, and the effect of variant mutations to be interpreted and predicted. Key to this is a protocol for the reproducible production and crystallization of the VHH-RBD complexes. Based on our experience, we describe a workflow for expressing and purifying the proteins, and the screening conditions for generating diffraction quality crystals of VHH-RBD complexes. Production and crystallization of protein complexes takes approximately twelve days, from construction of vectors to harvesting and freezing crystals for data collection.
Keywords: SARS CoV-2Background
Camelids (llamas, alpacas, and camels) produce a unique type of heavy chain only antibody that comprises variable domains (VHH) linked to Fc constant domains (CH2 and CH3). The VHHs can be produced as single domain antigen-binding proteins or nanobodies (Muyldermans, 2013), with wide applications in the life sciences (Pleiner et al., 2015; Jovcevska and Muyldermans, 2020). In response to the COVID-19 pandemic (Dhama et al., 2020), we and many other groups have developed VHH reagents that bind to the receptor binding domain (RBD) of the SARS-CoV-2 spike protein that block binding to ACE-2 at the cell surface, which is the primary interaction that leads to cell infection (Xiang et al., 2020; Hanke et al., 2020; Schoof et al., 2020; Koenig et al., 2021; Huo et al., 2020, 2021). Multimeric versions of these VHHs, either as trimers or IgG Fc fusions, have been shown to neutralise SARS-CoV-2 both in vitro and in animal models of COVID-19 (Huo et al., 2021; Nambulli et al., 2021). Structural analysis of VHH-RBD complexes has revealed two regions where epitopes are clustered, one at or close to the ACE-2 binding surface (cluster 2), and one at the opposite side of the RBD (cluster 1) (Tang et al., 2021). Information about the VHH binding sites has enabled the effect of mutations in the spike protein to be interpreted and predicted. In our experience, forming RBD complexes with two VHH that bind to orthogonal sites has been necessary for the crystallization of some VHH-RBD complexes (Huo et al., 2021).
VHHs are routinely produced in the E. coli strain WK6 as hexahistidine tagged proteins, using Isopropyl β-D-1-thiogalactopyranoside (ITPG) inducible promoters, and the addition of a secretion signal (e.g., pelB or ompA), to enable recovery of the VHH from the periplasm following induced expression. Primary purification uses immobilised metal affinity chromatography (IMAC) with gel filtration added as a polishing step. Alternatively, VHHs can be directly purified from lysed cells though, in our experience, periplasmic extraction by osmotic shock gives higher protein yields.
As a glycosylated protein, the production of the RBD of the SARS-CoV-2 spike protein requires expression in higher eukaryotic cells for which both insect and mammalian cells have been used. An appropriate signal sequence is added to the N-terminus to direct product to the cell media, and a hexahistidine tag added to the C-terminus for purification by IMAC. The N-glycosylation of the RBD (Asn331 and Asn343) introduces chemical heterogeneity, particularly if produced in mammalian cells, which generally inhibits crystallization (Chang et al., 2007). By growing cells in the presence of the mannosidase inhibitor kifunensine, N-glycosylation can be arrested at a high mannose state (GlcNAc2Man9 glycoforms). These can subsequently be trimmed back to single N-acetylglucosamine residues, by treatment with endoglycosidase F1 or H (Chang et al., 2007). Based on past experience (Nettleship et al., 2013), this is most efficiently carried out prior to purification of the VHH-RBD complex.
An issue for the purification of secreted glycoproteins from cell culture media by IMAC is that some media components displace the His-tagged protein-of-interest during the chromatography step, significantly reducing the yield. Here, we have used an automated method of affinity purification by IMAC and gel filtration, which involves loading the sample onto the Ni-NTA column in batches, with a column washing step between each batch (Nettleship et al., 2009).
We have reported the crystallization of a number of VHH-RBD complexes using commercially available screens, in 96-well format, and low sample volumes of 1–200 nL. Diffraction data were collected from the primary crystal hits using synchrotron X-rays, and structures solved by molecular replacement (Huo et al., 2020, 2021). Key to achieving good quality crystals was the preparation of the proteins. Thus, in this article, we describe our optimised workflow for producing VHH-RBD complexes and their crystallization for analysis by X-ray crystallography.
Materials and Reagents
Production of a receptor binding domain
Vector construction
pOPINTTG vector digested with KpnI/PmeI (Figure 1A)
Human codon optimised synthetic RBD gene (encoding amino acids 330–352) with 15-bp extensions overlapping with pOPINTTG in-fusion entry sites (lower case):
5’gcgtagctgaaaccggcCCGAATATCACAAATCTTTGTCCTTTCGGAGAAGTATTCAACGCAACTCGCTTCGCATCAGTATATGCCTGGAACCGCAAACGAATTTCTAATTGTGTCGCCGACTACTCTGTGCTTTACAACTCAGCATCATTTTCAACATTCAAATGCTATGGTGTCTCCCCTACGAAGTTGAACGATCTTTGTTTCACTAATGTCTACGCCGATTCCTTTGTTATTAGGGGGGACGAGGTTCGCCAGATCGCGCCGGGCCAAACGGGCAAGATAGCTGACTATAATTATAAGCTGCCAGACGACTTTACAGGCTGCGTTATCGCTTGGAATTCAAACAATCTTGATAGCAAAGTGGGTGGTAATTATAACTACCTCTACAGACTGTTTCGCAAGTCTAATCTGAAGCCTTTCGAGCGGGACATCTCTACTGAGATCTATCAAGCTGGTTCAACCCCCTGCAATGGCGTCGAGGGTTTTAACTGTTACTTCCCACTTCAGTCATACGGATTTCAACCAACTAATGGGGTTGGCTATCAGCCGTACCGCGTGGTCGTTCTTAGCTTTGAGCTGCTTCACGCCCCTGCAACGGTGTGCGGACCGAAAAAAAGTACAAAaaacatcaccatcac 3’
ClonExpress II One Step Cloning kit (Vazyme, catalog number: C113-02)
NucleoSpin 148® Gel and PCR Clean-up kit (MACHEREY-NAGEL, catalog number: 12303368)
Stellar competent cells (Takara Bio, catalog number: 636766)
LB medium (Sigma-Aldrich, catalog number: L3022)
S.O.C. (Super Optimal broth with Catabolite repression) recovery medium (ThermoFisher Scientific, catalog number: 15544034)
Ampicillin (Sigma-Aldrich, catalog number: A9393)
LB-agar plates supplemented with 100 μg/mL ampicillin
Plasmid miniprep kit (e.g., Qiagen, catalog number: 27104)
Plasmid Plus Midi kit (e.g., Qiagen, catalog number: 12941)
Sequencing primers: pTTfwd 5’ TCCACAGGTGTCCACTCC 3’
pTTrev 5’ TCCTTTATTAGCCAGAGG 3’
Expression in expi293TM cell
500 mL sterile baffled flasks with vented closure (ThermoFisher Scientific, catalog number: 4116-0500)
CountessTM cell counting chamber slides (ThermoFisher Scientific, catalog number: C10228)
Expi293TM cells (ThermoFisher Scientific, catalog number: A14527)
Expi293TM expression medium (ThermoFisher Scientific, catalog number: A1435101)
Trypan blue solution (ThermoFisher Scientific, catalog number: 15250061)
Hanks' Balanced Salt Solution (HBSS) (10×) (ThermoFisher Scientific, catalog number: 14185052)
Gibco OPTI-MEM reduced serum medium (ThermoFisher Scientific, catalog number: 31985062)
Valproic acid (Sigma-Aldrich, catalog number: P4543)
Glucose (45% solution) (see Recipes or from Sigma-Aldrich, catalog number: G8769)
Sodium propionate (Sigma-Aldrich, catalog number: P1880)
PEI MAX 40 K (Polysciences Inc., catalog number: 24,765-1)
Kifunensine (Sigma-Aldrich, catalog number: K1140)
Pre-cast SDS polyacrylamide gels (e.g., NuPAGETM 10%, Bis-Tris ThermoFisher Scientific, catalog number WG1201A)
InstantBlue® Coomassie protein stain (Abcam, catalog number: ab119211)
RBD purification
5 ml HisTrap_FF Ni-NTA columns (Cytiva, catalog number: 1752860)
SD75 16/600 size exclusion column (Cytiva, catalog number: 28989333)
Imidazole (Sigma-Aldrich, catalog number: I2399)
PBS, Phosphate Buffered Saline, 10× Solution (Fisher, catalog number: BP399-20)
Wash buffer (see Recipes)
Elution buffer (see Recipes)
Gel filtration buffers (see Recipes)
VHH production
Vector construction
pADL-23c vector cut with SfiI (Figure 1B) (New England Biolabs, catalog number: R0123S)
Infusion primers:
VHH Fwd primer 5’ GTTATTACTCGCGGCCCAGCCGGCCATGGCCC 3’
VHH Rev primer 5’ GGTGATGGTGTTGGCCTTTATTAATGATGGTGGTGATGGTG 3’
Proof reading polymerase (e.g., PhusionFlashTM high fidelity polymerase ThermoFisher Scientific, catalog number: F548S)
pADL-23c sequencing primers: PhDseqFwd 5’ GCTTCCGGCTCGTATGTTG 3’
PhDseqRev 5’ GTCGTCTTTCCAGACGTTAG 3’
2 mL Cryovials (e.g., ThermoFisher Scientific, catalog number: 5000-0020)
E. coli expression
WK6 cells Escherichia coli (Migula) Castellani and Chalmers (ATCC, catalog number: 47078)
Terrific Broth (TB) Medium (see Recipes)
Isopropyl β-D-1-thiogalactopyranoside, IPTG (Sigma-Aldrich, catalog number: I6758)
Glucose
Magnesium chloride
Ampicillin
VHH purification
TES buffer (see Recipes)
DNase I (Sigma-Aldrich, catalog number: D4263)
Magnesium Chloride (Sigma-Aldrich, catalog number: M8266)
Sucrose (Sigma-Aldrich, catalog number: S0389)

Figure 1. Vector maps (A) pOPINTTGneo (B) pADL-23c
Crystallization of VHH-RBD complexes
Preparation of VHH-RBD complexes
EndoH ( New England Biolabs, catalog number: P0702S)
SD200 10/300 size exclusion column (Cytiva, catalog number: 28990944)
Crystallization screening
Swisssci Triple-Drop crystallization plates (Molecular Dimensions, catalog number: MD11-003-100)
V well Microplate without lid, natural, polypropylene (Greiner, catalog number: 651201 96)
VIEWsealTM plate sealer, transparent, non-piercable Greiner, catalog number: 676070)
Pact premierTM crystallization screen condition: (Molecular Dimension, catalog number: MDSR-29)
JCSG-plusTM crystallization screen condition: (Molecular Dimensions, catalog number: MDSR-37)
SG1TM crystallization condition: (Molecular Dimensions, catalog number: MDSR-88)
Cryopreservation of crystals
Glycerol (Molecular Dimensions, catalog number: MD2-100-65)
PEG 400 (Sigma-Aldrich, catalog number: 06855)
Mounted Round LithoLoops (0.25 mm) (Molecular Dimensions, catalog number: MD7-137)
Mounted Round LithoLoops (0.15 mm) (Molecular Dimensions catalog number: MD7-135)
Standard Foam Dewar (Molecular Dimensions, catalog number: MD7-35)
Uni-Puck 10 Pack ( Molecular Dimensions, catalog number: MD7-613)
Cryotool set (Molecular Dimensions, catalog number: MD7-517)
Dry Shipper (Molecular Dimensions, catalog number: MD7-21
Equipment
Microvolume spectrophotometer (e.g., ThermoFisher Scientific, NanoDropTM One/OneC Microvolume UV-Vis Spectrophotometer: ND-ONE-W)
CO2 orbital shaker (e.g., n-Biotek.com ANICELL incubator, catalog number: NB-206CXL/NB)
Tabletop centrifuge suitable for 50 mL Falcon tubes (Sorvall Legend RT Plus)
Cell Counter (e.g., ThermoFisher Scientific CountessTM 3 Automated Cell Counter)
ÄKTA Xpress automated multi-step purification system
Liquid handler (e.g., Hydra Dispenser by Art Robbins Instruments)
Low volume dispenser for crystallization plates (e.g., SPT labtech mosquito® LV)
Crystal plate imager (e.g., Formulatrix RockImager® system)
Procedure
Production of receptor binding domains
Vector construction
For 10 μL of in-fusion reaction, mix 20 ng (1–3 μL) of synthetic gene, 100 ng (1–2 μL) of PmeI/NcoI cut pOPINTTG vector, 1 μL of Exnase II, and 2 μL of optimized buffer supplied with the cloning kit.
Incubate the reaction at 37°C for 30 min, and then stop immediately, by adding 20 μL of ice-cold TE buffer.
Use 5 μL of the resulting reaction mixture to transform 20 μL of Stellar competent cells, by incubating on ice for 30 min, then heat shocking at 42°C for 45 s.
Add 400 μL of SOC media and incubate at 37°C for 45 min.
Plate 100 μL of the cell and media mixture on LB-agar plates supplemented with 100 μg/mL ampicillin, and culture at 37°C overnight.
Pick single colonies into 3 mL of LB containing 100 μg/mL ampicillin and incubate at 37°C overnight.
Produce glycerol stocks, by adding 0.5 mL of 50% (v/v) sterile glycerol to 0.5 mL of overnight culture of cells in LB in a 2-mL cryovial, and freezing at -80°C.
Prepare DNA from pelleted cells, and confirm correct clones by sequencing minipreps DNAs with pTTfwd and PTTrev primers.
Use 50% glycerol stock to grow larger scale cultures for transfection-grade plasmid preparation.
Purify transfection-grade plasmids (0.5–1 mg) from 150 mL of overnight LB culture, using QIAGEN Plasmid Plus Midi kit and, if required, store plasmids at -20°C in sterile 1.5-mL Eppendorf tubes.
Measure the purity and concentration of DNA using NanoDrop spectrophotometer. Plasmid DNA used for transfections should be of high purity (see Note 1).
The concentration and purity of DNA is calculated automatically by the instrument using the following equations:
Concentration: DNA (µg/mL) = (A260 reading – A320 reading) × dilution factor × 50 µg/mL
Purity: (A260/A280) = (A260 reading – A320 reading) ÷ (A280 reading – A320 reading)
Use 1 μg DNA per one million of transfected cells.
Transfection protocol
Maintain the suspension culture of Expi293TM cells for at least three passages after defrosting (passage numbers 3–30 can be used in experiments) in a humidified (80%) incubator, with 5–8% CO2, at 37°C, on an orbital shaker at 120 rpm in Gibco Expi293TM Express medium, at a cell density between 0.5 and 5.0 × 106 cells/mL. Use a 125-mL flask to maintain 30 mL of Expi293 cells.
One day before transfection, seed Expi293TM cells at a cell density of 1 × 106 cells/mL. For each preparation of RBD, set up three cultures of 170 mL in 500-mL flasks.
On the day of transfection, determine cell count and viability, and if cell count is between 2 × 106 –2.5 × 106/mL and at least 95% viable, proceed with transfection (see Note 2).
For each 170-mL culture, mix 17 mL of OPTI-MEM media with 170 μg plasmid DNA and 918 μL of PEI Max 40kDa transfection reagent in a 50-mL Falcon tube.
Mix thoroughly, incubate at room temperature (RT) for 10 min, and add gently (dropwise) to the Expi293TM cells (see Note 3).
Add kifunensine from a stock concentration of 1 mg/mL (100 μL per 100 mL of culture volume), and return cells to incubate on the orbital shaker at 125 rpm, 5–8% CO2, 80% humidity and 37°C.
After 16–18 h, add to each 170 mL culture, 2,890 μL of valproic acid, 1,100 μL of sodium propionate, and 3,400 μL of glucose. Return the culture to the incubator (see Note 4).
On day 5 after transfection, determine cell count and viability.
Harvest media, and spin in a 0.5-L centrifuge bottle at 6,000 × g for 20 min. Filter sterilise using a 0.45 μm 0.5-L bottle top filter.
Purification
The IMAC-SEC purification protocol is for an ÄKTA Xpress platform using a programme described inNettleship et al. (2009). The same workflow can be implemented on other purification systems with automated peak detection. A transcript of the Unicorn programme is provided as an Appendix.
Equilibrate the gel filtration column using the “Gel Filtration Equilibration” programme and the Gel filtration buffer (20 mM Tris, pH 7.5, 200 mM NaCl).
Follow the method “Mammalian Prepping System” to set up the system. It will pump wash A1 and A2, and clean the inlets. It will then ask you to screw the 5-mL column into position 1, and will equilibrate the column.
Add an equal volume of PBS to the filtered cell supernatant, and adjust the pH to 7.4 with NaOH.
Once the column is plugged in and the sample is ready to be loaded, slowly remove the A2 line from the buffer (by small movements to avoid bubbles in the line), and insert it into the sample. Make sure all the lines are at the bottom of the bottles.
Leave the protocol to run overnight.
Using the trace from the ÄKTA, select the correct fractions containing the protein of interest. Mix 10 μL of protein solution with 10 μL of sample buffer, and heat at 95°C for 5 min. Run an SDS-PAGE gel of the samples, and stain it using Instant Blue®.
Concentrate the protein, usually to 5 mg/mL, using a 10 kDa concentrator centrifuge at 2,500 × g and 4°C.
Aliquot (e.g., 0.1 mL) and flash freeze the protein, by plunging tubes into liquid nitrogen.
Store at -80°C.
Production of VHHs
Vector construction
VHHs identified by screening M13 phage display libraries are re-expressed for protein production.
Amplify VHH from source display vector using VHH Fwd primer and VHH Rev primers and the following PCR conditions:
1) 98°C for 10 s
2) 30 cycles of:
98°C for 1 s
60°C for 5 s
72°C for 15 s
3) 72°C for 2 min
4) 4°C hold
Purify amplified VHH DNA by agarose gel electrophoresis, and extract using the Nucleospin® kit according to the manufacturer’s instructions. Clone PCR product into the SfiI cut pADL-23c vector, as described for the cloning of the RBD.
Verify clones, by sequencing with PhDseqFwd and PhDseqRev primers.
Expression in E. coli
Transform chemically competent WK6 cells as described above, for construction of the RBD expression vector.
Pick a colony from the plate of transformed cells, and set up a pre-culture: 8 mL of TB supplemented with 100 μg/mL ampicillin, 2% glucose and 1 mM MgCl2.
Set up the culture: 800 mL of TB medium in a 2-L flask supplemented with 100 μg/mL ampicillin, 0.1% glucose, and 1 mM MgCl2.
Pre-warm the flask/medium to 37°C.
Add pre-culture and grow culture on an orbital shaker at 225 rpm and 37°C.
Add IPTG to a final concentration of 1 mM when OD600 reaches ~1.2 (usually takes around 3.5 h), and continue growing at 225 rpm and 28°C overnight.
Pellet the cells at 2,500 × g at 4°C for 15 min.
Purification of VHHs
Add 15 mL of TES buffer to the cell pellet, and resuspend slowly in a bottle with a magnetic stirrer at 4°C overnight.
The next day, add twice the volume of cold TES/4 buffer supplemented with 120U Kunitz DNaseI to the resuspended culture, and slowly stir for 2 h.
Top up to 80-mL with TES/4 buffer.
Spin down the pellet at 28,000 × g and 4°C for 30 min (in a 50-mL tube).
Filter the supernatant through a 0.8 μm filter under vacuum.
Dilute the supernatant with five volumes of PBS (pH 7.4), and mix well.
Load the sample onto two 5-mL IMAC columns connected in series, at <2 mL/min.
Wash the column with PBS (pH 7.4) containing 30 mM imidazole.
Elute the sample with PBS containing 300 mM imidazole at 1 mL/min into a 96-well collection plate, collecting 1-mL fractions.
Pool A280 peak fractions (approximately 7.5 mL) and run on a Superdex S75 16/600 in gel filtration buffer (50 mM Tris pH 7, 150 mM NaCl).
Pool A280 peak protein fractions, and concentrate using a 5 kDa MWCO concentrator, usually to 15 mg/mL (see Note 5).
Aliquot (e.g., 0.1 mL) and flash freeze the protein, by plunging tubes into liquid nitrogen.
Store at -80°C.
Crystallization of VHH-RBD complexes
Preparation of VHH:RBD complexes
Mix 5 mg of RBD (5 mg/mL) with 3 mg VHH (15 mg/mL) at a molar ratio RBD:VHH of 1:1.2, and incubate under agitation at 2 rpm in a cold room for 3 h (see Note 6).
Incubate the RBD-VHH complex with 0.4 mg of EndoH glycosidase (1 mg/mL) under agitation at 2 rpm at room temperature overnight (see Note 7).
Concentrate the mixture to 1 mL with a 5 kDa MWCO concentrator, and inject onto a Superdex 200 10/300 in gel filtration buffer (50 mM Tris pH 7, 150 mM NaCl).
Monitor A280, pool peak fractions, and concentrate using 5 kDa MWCO concentrator to 20 mg/mL (see Figures 2 and 3).
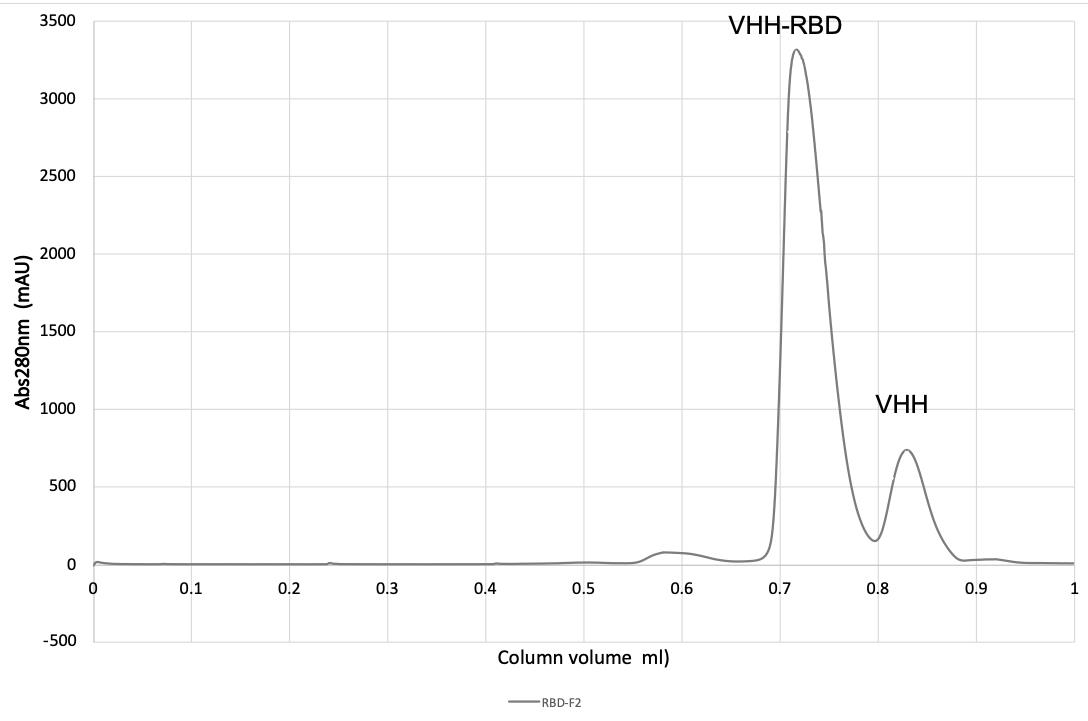
Figure 2. Purification of VHH-RBD complex on a SD200 20/300 size exclusion column (CV 23.56 mL).
Figure 3. SDS-Page of RBD (lane 1), VHH (lane 2), and fractions from gel filtration of RBD-VHH complexes, following treatment with endoglycosidase HH (lanes 3–12).Crystallization screening
Dispense 25 μL of crystallization solution from 96-well masterblock into the reservoir well of Swisssci crystal plate, using the Hydra liquid handler.
Pipette the protein solution into a column in a V well microplate well plate.
Set up a sitting drop plate comprising 288 drops, using the robotic liquid handler (Mosquito) using three protein:reservoir ratios (100 nL protein + 100 nL reservoir; 200 nL protein + 100 nL reservoir; 100 nL protein + 200 nL reservoir) (see Note 8).
Seal completed plate using VIEWseal.
Image and store plates by Fibonacci schedule in Formulatrix imager at 20°C (see Figure 4 for examples of crystals).
Cut out a square of seal over the selected crystal drop, and open drop.
Add 1 μL of cryoprotectant mix (30% glycerol or Peg 400 in original crystallization condition) onto crystals/drop. Table 1 gives examples of crystallized VHH-RBD complexes.
Place mounting pin onto magnetic wand, loop crystal from drop, and place rapidly into foam dewar containing liquid nitrogen.
Store mounted crystal in puck within foam dewar.
Transfer puck into liquid nitrogen pre-cooled Dry shipper.
Table 1. Examples of crystallized VHH-RBD complexes with details of crystallization conditions, the space group, and resolution limit of crystals generated.
Complex RBD-F2 RBD-H3-C1 RBD-C5 Crystal screen PactTM JCSGTM SG1TM Crystallisation condition 0.1 M SPG, pH 8, 25% Peg 1500 1.0 M Lithium chloride, 0.1 M Citrate pH 4, 20% Peg 6000 0.2 M Na Acetate, 0.1 M Na Cacodylate pH 6.5 and 30% w/v PEG 8000 Time for crystal formation 3 days less than 24 h 3 days optimal growth Ratio 0.2 μL protein with 0.1 μL reservoir 0.2 μL protein with 0.1 μL reservoir 0.1 μL protein with 0.1 μL reservoir Concentration 34 mg/mL 18 mg/mL 18 mg/mL Cryoprotectant glycerol Peg400 glycerol Data collection Exposure 0.006s 0.012s 0.008s Space group P31 P41212 P21212 Resolution (Å) 2.3 1.9 1.5

Figure 4. Crystal shape of the different complexes: RBD-F2 (left), RBD-H3-C1 (middle), and RBD-C5 (right).
Notes
Good quality DNA with minimum protein and chemical contamination should have ratios of absorbance 260/280 between 1.8–2.0, and 260/230 between 2.0–2.2.
Our protocol is suitable for any scale of expression: 1–3 mL plate experiments, and 30–300 mL up-scaled expression in flasks. Scale provided volumes and quantities of reagents proportionally to the volume of transfected cells.
Do not mix DNA and PEI directly, as they will precipitate immediately.
The use of valproic acid, sodium propionate (histone deactylase inhibitors), and glucose feed in combination helps substantially enhance gene expression.
It is important to check periodically that there is no precipitation of the VHH during the concentration step and to stop further concentrating the protein if this is observed. Any precipitate can be removed by centrifugation of the sample in a microfuge at 12,000 × g for 10 min and then measuring the final protein concentration by absorbance at 280 nm.
A molar excess of VHH is added, to ensure that the RBD is fully complexed with the nanobody.
EndoH has maximum efficiency at pH ~5.2. However, as the majority of proteins are unstable at this pH, the reaction is carried out at pH 7.5. Also, many proteins are unstable at 37°C for long periods of time, so RT is preferred for the reaction. EndoH is active at RT and at pH 7.5, but the reaction takes longer than it would if it was run under optimal conditions.
The two sparse matrix screens (JCSG+ and SG1) were routinely screened at two different concentrations (usually, 34 mg/mL, and 18 mg/mL) with three different ratios. For the complexes RBD-F2 and RBD-H3-C1, we obtained a number of crystallization hits, while for RBD-C5 we only obtained one hit. Only occasionally the PACT screen was used. Crystals were harvested straight from the sparse matrix screen and did not require further optimization. To obtain more crystals with very subtle different diffraction quality, usually the same condition was set up as a row on the mosquito, using the same protocol. This also allowed testing of two different cryo-conditions (usually, glycerol, and Peg 400). Crystals were very reproducible. The cleanest crystals were harvested from all hits usually within 24 h of growth, but diffraction quality remained stable up to 5 days thereafter for most hits.
Recipes
Valproic acid
500 mg in 10 mL of cell media or HBSS, filter 0.2 μm, and store at -20°C.
Propionate solution
1 g sodium propionate in 10 mL of cell media or HBSS, filter, and store at -20°C.
PEI
Suspend 100 mg of PEI Max 40K in 90 mL of MilliQ water. Stir using a PTFE coated stirring bar. It should take less than 5 min to dissolve, then adjust the pH using NaOH or HCl to 7.0. Add water to 100 mL, filter 0.2 μm, and store at -20°C.
Glucose
45 g in 100 mL of HBSS or cell medium, filter 0.2 μm, and store at -20°C.
Wash buffer
PBS + 30 mM imidazole pH 7.4
Elution buffer
PBS + 300 mM imidazole pH 7.4
Gel filtration buffers
20 mM Tris, and pH 7.5, 200 mM NaCl, or 50 mM Tris pH 7, 150 mM NaCl
TES buffer
200 mM Tris pH 8, 0.5 mM EDTA, 500 mM sucrose
TES/4 buffer
50 M Tris pH 8, 125 mM sucrose
TB medium
Yeast extract (24 g/L), Tryptone (20 g/L), Glycerol (4 mL/L), Phosphate buffer (100 mL/L) of 0.17 M KH2PO4, 0.72 M K2HPO4
Acknowledgments
This work was supported by the Rosalind Franklin Institute, funding delivery partner EPSRC. and the Rosalind Franklin Institute EPSRC grant no. EP/ S025243/1. J.H.N., A.L.B. are supported by Wellcome Trust (100209/Z/12/Z). J.H. is supported by the EPA Cephalosporin and Edward Penley Abraham Funds. X-ray data were obtained using Diamond Light Source COVID-19 Rapid Access time on Beamline I03, I04 and I24 (proposal MX27031).
Competing interests
The Rosalind Franklin Institute has filed a patent on the identification of nanobodies to the spike protein of SARS-CoV-2; R.J.O., J.H. and J.H.N. are named as inventors. The other authors declare no competing interest.
References
- Chang, V. T., Crispin, M., Aricescu, A. R., Harvey, D. J., Nettleship, J. E., Fennelly, J. A., Yu, C., Boles, K. S., Evans, E. J., Stuart, D. I., et al. (2007). Glycoprotein structural genomics: solving the glycosylation problem. Structure 15(3): 267-273.
- Dhama, K., Khan, S., Tiwari, R., Sircar, S., Bhat, S., Malik, Y. S., Singh, K. P., Chaicumpa, W., Bonilla-Aldana, D. K. and Rodriguez-Morales, A. J. (2020). Coronavirus Disease 2019-COVID-19. Clin Microbiol Rev 33(4): e00028-20.
- Hanke, L., Vidakovics Perez, L., Sheward, D. J., Das, H., Schulte, T., Moliner-Morro, A., Corcoran, M., Achour, A., Karlsson Hedestam, G. B., Hallberg, B. M., et al. (2020). An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat Commun 11(1): 4420.
- Huo, J., Le Bas, A., Ruza, R. R., Duyvesteyn, H. M. E., Mikolajek, H., Malinauskas, T., Tan, T. K., Rijal, P., Dumoux, M., Ward, P. N., et al. (2020). Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat Struct Mol Biol 27(9): 846-854.
- Huo, J., Mikolajek, H., Le Bas, A., Clark, J. J., Sharma, P., Kipar, A., Dormon, J., Norman, C., Weckener, M., Clare, D. K., et al. (2021). A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19. Nat Commun 12(1): 5469.
- Jovcevska, I. and Muyldermans, S. (2020). The Therapeutic Potential of Nanobodies. BioDrugs 34(1): 11-26.
- Koenig, P. A., Das, H., Liu, H., Kummerer, B. M., Gohr, F. N., Jenster, L. M., Schiffelers, L. D. J., Tesfamariam, Y. M., Uchima, M., Wuerth, J. D., et al. (2021). Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape. Science 371(6530): eabe6230.
- Muyldermans, S. (2013). Nanobodies: natural single-domain antibodies.Annu Rev Biochem 82: 775-797.
- Nambulli, S., Xiang, Y., Tilston-Lunel, N. L., Rennick, L. J., Sang, Z., Klimstra, W. B., Reed, D. S., Crossland, N. A., Shi, Y. and Duprex, W. P. (2021). Inhalable Nanobody (PiN-21) prevents and treats SARS-CoV-2 infections in Syrian hamsters at ultra-low doses. Sci Adv 7(22): eabh0319.
- Nettleship, J. E., Rahman-Huq, N. and Owens, R. J. (2009). The production of glycoproteins by transient expression in Mammalian cells. Methods Mol Biol 498: 245-263.
- Nettleship, J. E., Ren, J., Scott, D. J., Rahman, N., Hatherley, D., Zhao, Y., Stuart, D. I., Barclay, A. N. and Owens, R. J. (2013). Crystal structure of signal regulatory protein gamma (SIRPgamma) in complex with an antibody Fab fragment. BMC Struct Biol 13: 13.
- Pleiner, T., Bates, M., Trakhanov, S., Lee, C. T., Schliep, J. E., Chug, H., Bohning, M., Stark, H., Urlaub, H. and Gorlich, D. (2015). Nanobodies: site-specific labeling for super-resolution imaging, rapid epitope-mapping and native protein complex isolation. Elife 4: e11349.
- Schoof, M., Faust, B., Saunders, R. A., Sangwan, S., Rezelj, V., Hoppe, N., Boone, M., Billesbolle, C. B., Puchades, C., Azumaya, C. M., et al. (2020). An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science 370(6523): 1473-1479.
- Tang, Q., Owens, R. J. and Naismith, J. H. (2021). Structural Biology of Nanobodies against the Spike Protein of SARS-CoV-2. Viruses 13(11): 2214.
- Wec, A. Z., Wrapp, D., Herbert, A. S., Maurer, D. P., Haslwanter, D., Sakharkar, M., Jangra, R. K., Dieterle, M. E., Lilov, A., Huang, D., et al. (2020). Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science 369(6504): 731-736.
- Xiang, Y., Nambulli, S., Xiao, Z., Liu, H., Sang, Z., Duprex, W. P., Schneidman-Duhovny, D., Zhang, C. and Shi, Y. (2020). Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2. Science 370(6523): 1479-1484.
Article Information
Copyright
© 2022 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Le Bas, A., Mikolajek, H., Huo, J., Norman, C., Dormon, J., Naismith, J. H. and Owens, R. J. (2022). Production and Crystallization of Nanobodies in Complex with the Receptor Binding Domain of the SARS-CoV-2 Spike Protein. Bio-protocol 12(9): e4406. DOI: 10.21769/BioProtoc.4406.
Category
Biophysics > X-ray crystallography
Biochemistry > Protein > Expression
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








