- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Yeast Lipid Extraction and Analysis by HPTLC
Published: Vol 11, Iss 13, Jul 5, 2021 DOI: 10.21769/BioProtoc.4081 Views: 5389
Reviewed by: Alexandros AlexandratosAnu P. MinhasAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Yeast Single-cell RNA-seq, Cell by Cell and Step by Step
Mariona Nadal-Ribelles [...] Lars M. Steinmetz
Sep 5, 2019 7713 Views

A Microfluidic Platform for Tracking Individual Cell Dynamics during an Unperturbed Nutrients Exhaustion
Théo Aspert [...] Gilles Charvin
Jul 20, 2022 2889 Views

Analysis of Lipid-linked Oligosaccharides Synthesized in vivo in Schizosaccharomyces pombe
Ayelen Valko [...] Cecilia D’Alessio
Sep 20, 2022 2227 Views
Abstract
The diversity of lipid structures, properties, and combinations in biological tissues makes their extraction and analysis an experimental challenge. Accordingly, even for one of the simplest single-cellular fungi, the budding yeast (Saccharomyces cerevisiae), numerous extraction and analysis protocols have been developed to separate and quantitate the different molecular lipid species. Among them, most are quite sophisticated and tricky to follow. Herein, we describe a yeast total lipids extraction procedure with a relatively good yield, which is appropriate for subsequent thin-layer chromatography (TLC) or liquid chromatography-mass (LC-MS) analysis. We then discuss the most widely used solvent systems to separate yeast phospholipids and neutral lipids by TLC. Finally, we describe an easy and rapid method for silica gel staining by a Coomassie Brilliant Blue-methanol mixture. The stained lipid species can then be quantitated using imaging software such as ImageJ. Overall, the methods described in this protocol are time-saving and novice-friendly.
Keywords: Lipid extractionBackground
Lipid research has gained great momentum in recent years, stimulated in part by the interest in lipid-associated disorders in humans, in part by aspirations of biofuel and biopharmaceutical production. Lipids play vital roles in many biological processes, including the formation of bio-membranes, the storage of energy, and numerous signal transduction processes. The budding yeast, Saccharomyces cerevisiae, has been an excellent model organism for studying the physiological and pathophysiological roles of lipids at the cellular level (Popa et al., 2016). Primary lipid metabolism is highly conserved between yeast and humans in both the basic biochemical pathways and the regulatory circuitries that link lipid metabolism to energy homeostasis, cell growth, and development. One advantage of using yeast in lipid-related research is that the pathways and molecular compositions are often less complicated than those in mammals, facilitating their scientific investigation. In yeast, PA (phosphatidate acid), PC (phosphatidylcholine), PE (phosphatidylethanolamine), PI (phosphatidylinositol), PS (phosphatidylserine), DAG (diacylycerols), and their respective lyso-derivatives are the most abundant membrane lipid classes, while PG (phosphatidylglycerol) and CL (cardiolipin) are less abundant. TAG (triacylglycerol) and SE (sterol ester) act as storage lipids. The most abundant yeast sphingolipids consist of IPC (inositolphosphoceramide), MIPC (mannose inositolphosphoceramide), and M(IP)2C (mannose diinositolphosphoceramide) (Klug and Daum, 2014).
Lipid extraction procedures described in the literature generally share common principles but vary in processing steps. Among these, the most cited Folch method uses CHCl3/CH3OH (2:1 v/v) as the solvent system (Folch et al., 1957). Alternate proportions of CHCl3/CH3OH or replacement of CHCl3 with 2-propanol or hexane have also been utilized (Fuchs et al., 2015; Knittelfelder and Kohlwein, 2017a and 2017b). In this protocol, we use CHCl3/CH3OH (2:1 v/v) as the solvent for lipid extraction and break yeast cells with glass beads in a mechanical bead mill. We repeat the extraction five times to ensure efficient lipid extraction.
Silica gel is the most wildly used stationary phase for chromatographic lipid separation. Impregnated with different substances, modified silica gel can be used to separate various lipid classes. Among the most popular modifications, silver nitrate impregnation has better potential for separating glycerolipids containing unsaturated fatty acyl chains (Dobson et al., 1995); boric acid is primarily used to separate the various isomers of DAG or PL (Ando et al., 2000). Like in other chromatographic techniques, TLC can be classified as “normal” or “reversed” phase based on the polarities of their mobile and stationary phases. Normal phase (polar stationary phase and non-polar mobile phase) chromatography is the standard method to separate lipids of interest that may have different polarities caused by a variation in their head groups (Fuchs et al., 2011). Typically, the size of gel particles for TLC is 10-50 μm. In contrast, the size of the high-performance thin-layer chromatography (HPTLC) gel particles is about 5 μm with narrow distributions (Fuchs et al., 2011), which results in higher separation quality and smaller sample quantity. It is noteworthy that bio-lipids are very complex molecules, and no single separation method is sufficient to separate all lipid species. With the stationary phase chosen, the separating efficiency of TLC or HPTLC mainly relies on the mobile phase. A 0.4% (NH4)SO4-impregnated silica gel plate together with chloroform-methanol-acetic acid-acetone-water (40:25:7:4:2 v/v/v/v/v) as the mobile phase can be used to separate phospholipids, lysophospholipids, and SM (sphingomyelin) (Wang and Gustafson, 1992). A chloroform-methanol-water mixture (35:15:2 or 50:40:10 v/v/v) is generally used for the separation of SM; a different proportion, chloroform-methanol-water mixture (65:25:4 v/v/v), can be used to separate phospholipids (Knittelfelder and Kohlwein, 2017a and 2017b). A mobile phase consisting of hexane-diethyl ether-acetic acid (70/30/1 v/v/v) is initially used to separate cholesterol isomers; it can also be used to separate neutral lipids such as TAGs, DAGs, and MAGs (monoacylglycerols). Alternatively, neutral lipids can be separated by TLC silica gel 60 plates and petroleum ether-diethyl ether-acetic acid (32:8:0.8 v/v/v/) mixture as the mobile phase (Knittelfelder and Kohlwein, 2017a and 2017b). Overall, it is a trial-and-error process to choose a proper stationary phase and mobile phase based on the characteristics of individual lipid species. In the present protocol, a silica gel 60 plate is used as the stationary phase, and a hexane-diethyl ether-acetic acid (70/30/1 v/v/v) mixture and a chloroform-methanol-water mixture (65:25:4 v/v/v) are used as the mobile phase to separate neutral lipids and phospholipids, respectively. Note that the mobile phase for chromatographic lipid separation is not to be confused with the solvent system for lipid extraction, which is discussed in the preceding paragraph.
After separation, the resulting positions of different lipid species on TLC plates need to be visualized, usually by color reactions with chemical spray reagents. An iodine vapor bath is one of the most widely used methods. The brown iodine color will disappear spontaneously once the plates are removed from the iodine vapor (Palumbo and Zullo, 1987). It has been reported that iodine can be difficult to remove from highly unsaturated lipids, but in our experience, the brown color fades away too quickly before follow-up steps can be properly performed. Alternative spray reagents include 2,7-dichlorofluorescein, rhodamine 6 G, and primuline. Together with iodine, they belong to non-destructive reagents, with the modification of lipid structures being reversible (Fuchs et al., 2011). 0.2% amino black 10 B in 1 M NaCl and 3.2% H2SO4, 0.5% MnCl2 in 50% ethanol are two destructive reagents with high sensitivity (Fuchs et al., 2011 and 2015; Knittelfelder and Kohlwein, 2017a and 2017b). In this protocol, we use Coomassie Brilliant Blue R-250, a non-destructive reagent first used to stain lipids in 1984 (Nakamura and Handa, 1984). In our experience, the detection sensitivity and durability of Coomassie Brilliant Blue R-250 are better than those of iodine vapor. Conveniently, the staining steps only take 30 min.
Materials and Reagents
Pipette tips (Thermo Fisher Scientific, different sizes and types)
Centrifuge tube, 5-ml round-bottomed (Sangon Biotech, catalog number: F610888)
Cryogenic microtubes, 1.5-ml deep cap, conical-bottomed, sterile (Sangon Biotech, catalog number: F600154)
Gel blotting paper (Sangon Biotech, catalog number: F513323)
Yeast extracts (OXCID, catalog number: LP0021)
Peptone (BD, catalog number: 211677)
D-glucose (Sangon Biotech, catalog number: A501991)
Yeast nitrogen base without amino acids and ammonium sulfate (Sangon Biotech, BBI, catalog number: A600505)
Chloroform (General-Reagent, catalog number: G75915B)
Methanol (General-Reagent, catalog number: G75851B)
Coomassie Brilliant Blue R-250 (Sangon Biotech, catalog number: CB0037)
Hexane (General-Reagent, catalog number: G14153D)
Diethyl ether (HUSHI, catalog number: 10009318)
Acetic acid (General-Reagent, catalog number: G73562B)
Lipid standard: 18:1 DG (1,2-dioleoyl-sn-glycerol) 2 mg/ml chloroform (Avanti, catalog number: 800811C)
Lipid standard: Egg PC (L-alpha-phosphatidylcholine) 10 mg/ml chloroform (Avanti, catalog number: 840051C)
TLC Silica gel 60 (25 aluminium sheets 20 × 20cm; Merk Millipore, catalog number: 105553)
Glass thin-layer chromatography (TLC) developing chambers (100 × 100, generic)
Glass beads (40 or 50 mesh, generic)
Equipment
Pipettes (Thermo Fisher Scientific, different types)
Shaking incubator (CRYSTAL, model: IS-RDH1)
Spectrophotometer (Shanghai Jing-Hua, model: 722S)
Centrifuge (Thermo, model: SORVALL ST16R)
Freeze-dryer/lyophilizer (Labconco, model: Freezone 6 Plus)
Analytical scale (Shanghai Heng-Ping, model: FA2104)
Drying oven (BOXUN, model: GZX-9146MBE)
Bead mill (Shanghai Jing-Xin, model: Tissue lyser-96)
Thermomixer (ALLSHENG, model: MSC-100)
TLC developing chamber with cover (generic)
Gel imaging system (Bio-Rad, model: Gel Doc 721BR05189)
Software
ImageJ (https://imagej.net/Downloads)
Procedure
Cell growth and harvest
Grow yeast cells in 100 ml YPD medium (1% yeast extract, 2% peptone, 2% glucose) overnight in a 30°C shaking incubator (250 rpm) until the optical density (OD600) of the culture is about 0.7-1.0.
Harvest the cells by centrifugation, 10,000 × g for 5 min at room temperature. Remove the supernatant completely and discard.
Freeze-dry the cells in a freeze-dryer and store at -80°C.
Total lipid extraction
For each freeze-dried sample, transfer 0.25 mg into a 1.5-ml cryogenic microtube.
Add 200 μl washed glass beads to each sample tube.
Add 500 μl chloroform-methanol (2:1, v/v) to each sample tube. Screw on the Teflon-sealed cap tightly.
Place the sample tubes in a bead mill and perform mechanical shearing at maximum speed for 5 min at room temperature. Repeat the mechanical lysis twice; rearrange tube locations in-between to avoid uneven cell lysis (Step 1 in Figure 1).
Centrifuge the cell lysates at room temperature, 10,000 × g for 5 min. After centrifugation, cell debris will be sandwiched between the supernatant and the glass beads (Step 2 in Figure 1). Transfer the supernatant to new tubes for collection (Step 3 in Figure 1).
Repeat the chloroform-methanol extraction procedure (Steps 2-6 in Figure 1) five times.
Merge the supernatant collections of each sample in a 5-ml tube and evaporate on a compact thermomixer at 60°C overnight with the lid off. This step needs to be performed in a fume hood. After evaporation, membrane-like substances will be left at the bottom of the tubes.

Figure 1. Schematic depiction of the lipid extraction procedure
Lipid analysis by TLC
Dissolve the lipid extracts in 100-500 μl chloroform.
Prepare the solvent mixtures:
For separation of phospholipids, prepare a chloroform-methanol-H2O (65:25:4, v/v/v) mixture (Knittelfelder and Kohlwein, 2017a and 2017b).
For separation of neutral lipids, prepare a hexane-diethyl ether-acetic acid (70:30:1 v/v/v) mixture (Fuchs et al., 2015).
Add the mobile phase to glass TLC developing chambers. Saturate the chamber atmosphere with folded blotting paper for more than 30 min.
Activate the silica gel 60 plates at 100°C in an oven for 30 min. Draw a fine spotting line with a pencil 2 cm from the bottom of the TLC plate.
Spot 5-20 μl dissolved samples on the activated silica gel plates carefully with 10-μl pipette tips. Keep the spacing between these spots larger than 0.5 cm. In addition, spot 0.2-1 μg lipid standards (DAG or PC) to distinguish the complex bands of each sample and to monitor the developing and staining steps.
Develop each plate in a solvent chamber until the solvent front reaches the top of the plate. To maintain a solvent-saturated atmosphere in the chamber, do not open the lid while the plate is developing.
Dry the plates in a fume hood for 20 min at room temperature.
Stain the dried plates with a 0.03% Coomassie Brillant Blue R-250 solution containing 20% methanol for 15 min, and subsequently de-stain in 20% methanol for 10 min (Nakamura and Handa, 1984).
Dry the plates under room temperature for 1 h and take photos by a gel imaging system (Figure 2).
Analyze the grayscale of each lipid band using ImageJ (Steps: Open an image file in ImageJ – go to Analyze – gels – Select First Line – Select Next Line – Wand (tracing) Tool – Plot Lanes).
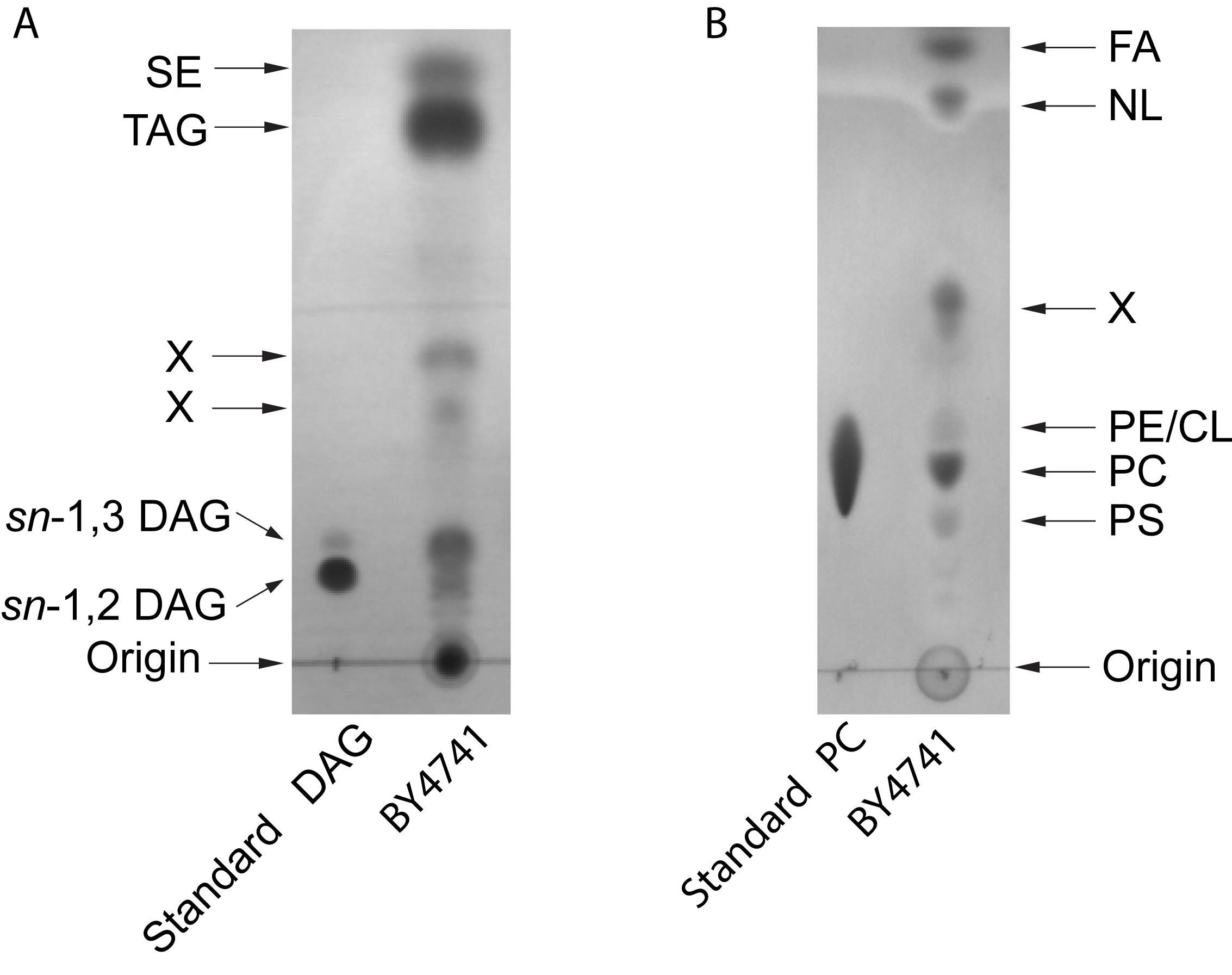
Figure 2. Examplary HPTLC separation of yeast lipids. A. Neutral lipids: the solvent system was hexane-diethyl ether-acetic acid (70:30:1 v/v/v). DAG, diacylglycerol; TAG, triacylglycerol; SE, steryl esters. B. Polar lipids: the solvent system was chloroform-methanol-H2O (65:25:4, v/v/v). FA, fatty acids; NL, neutral lipids; PE, phosphatidylethanolamine; CL, cardiolipin; PC, phosphatidylcholine; PS, phosphatidylserine. X, unidentified. Lipids were stained with Coomassie Brillant Blue R-250.
Acknowledgments
The authors would like to thank Dr. Zhou Peng and Prof. Xiaoling Miao (School of Life Sciences and Biotechnology, Shanghai Jiao Tong University) for their kind advice regarding experimental procedures. This work was supported by the National Natural Science Foundation of China (grants 91957104, 31671431, and 91754110). This protocol is adapted from Li et al. (2020) and works cited therein.
Competing interests
The authors declare no competing financial interests
References
- Ando, Y., Satake, M., and Takahashi., Y. (2000). Reinvestigation of positional distribution of fatty acids in docosahexaenoic acid-rich fish oil triacyl-sn-glycerols. Lipids 35(5): 579-582.
- Dobson, G., Christie, W.W., and Nikolova-Damyanova., B. (1995). Silver ion chromatography of lipids and fatty acids. J Chromatogr B Biomed Appl 671(1-2): 197-222.
- Folch, J., Lees, M. and Sloane Stanley, G. H. (1957). A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 226(1): 497-509.
- Fuchs, B., Süss, R., Teuber, K., Eibisch, M. and Schiller, J. (2011). Lipid analysis by thin-layer chromatography--a review of the current state. J Chromatogr A 1218 (19): 2754-2774.
- Fuchs, B., Popkova, Y., Süß, R. and Schiller, J. (2015). Separation of (Phospho)Lipids by Thin-Layer Chromatography. In: Instrumental Thin-Layer Chromatography. 2015: 375-405.
- Klug, L. and Daum, G. (2014). Yeast lipid metabolism at a glance. FEMS Yeast Res 14(3): 369-388.
- Knittelfelder, O. L. and Kohlwein. S. D. (2017a). Lipid Extraction from Yeast Cells. Cold Spring Harb Protoc 2017(5): pdb prot085449.
- Knittelfelder, O. L. and Kohlwein, S. D. (2017b). Thin-Layer Chromatography to Separate Phospholipids and Neutral Lipids from Yeast. Cold Spring Harb Protoc 2017(5).
- Nakamura, K. and Handa, S. (1984). Coomassie brilliant blue staining of lipids on thin-layer plates. Anal Biochem 142(2): 406-410.
- Popa C, Coll NS, Valls M and Sessa G. (2016). Yeast as a Heterologous Model System to Uncover Type III Effector Function. PLoS Pathog 12(2): e1005360.
- Palumbo, G. and Zullo, F. (1987). The use of iodine staining for the quantitative analysis of lipids separated by thin layer chromatography. Lipids 22(3): 201-205.
- Wang, W.Q. and Gustafson. A. (1992). One-dimensional thin-layer chromatographic separation of phospholipids and lysophospholipids from tissue lipid extracts. J Chromatogr 581(1): 139-142.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Li, D., Zhang, Z., He, C. and Xie, Z. (2021). Yeast Lipid Extraction and Analysis by HPTLC. Bio-protocol 11(13): e4081. DOI: 10.21769/BioProtoc.4081.
Category
Biochemistry > Lipid > Lipid isolation
Microbiology > Microbial biochemistry > Lipid
Microbiology > Microbial cell biology > Cell isolation and culture
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








