- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Isolation and CryoTEM of Phages Infecting Bacterial Wine Spoilers
Published: Vol 10, Iss 21, Nov 5, 2020 DOI: 10.21769/BioProtoc.3801 Views: 4411
Reviewed by: Alba BlesaRajesh ThippeshappaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A β-glucuronidase (GUS) Based Bacterial Competition Assay to Assess Fine Differences in Fitness during Plant Infection
Julien S. Luneau [...] Alice Boulanger
Jul 5, 2022 3219 Views

Tomato Stem Injection for the Precise Assessment of Ralstonia solanacearum Fitness in Planta
Yaru Wang [...] Alberto P. Macho
Aug 20, 2021 3721 Views

Isolation of Viral Biofilms From HTLV-1 Chronically Infected T Cells and Integrity Analysis
Coline Arone [...] Delphine Muriaux
Dec 20, 2024 1828 Views
Abstract
With the objective to isolate phages infecting wine bacterial spoilers, we designed a method for the isolation and purification of phages infecting grape-associated bacteria. The method proved successful to isolate GC1 tectivirus infecting the acetic acid bacterium Gluconobacter cerinus. The isolated phage represents a new genus within the Tectiviridae, named "Gammatectivirus". Using a traditional technique for the concentration of phage particles involving several steps of centrifugation, further insights in the ultrastructure of GC1 could be observed by cryo electron microscopy, saving time and effort. The simple workflow presented may be applied to other viruses infecting bacteria inhabiting other vegetal niches.
Graphic abstract
Flowchart illustrating the protocol to isolate, concentrate and observe GC1 under cryo-EM
Background
Winemaking is a complex and fluctuating environment that is characterized by the temporal succession of distinct communities of microorganisms. The wine-making process starts with the selection of the fruit and the fermentation of sugars into alcohol by yeasts. In most red and dry white wines, this step is followed by a malolactic fermentation, which reduces acidity, increases microbial stability, and creates good-quality grape wine. In contrast to other food fermentations, little knowledge is so far available on the diversity of viruses infecting bacteria, also known as bacteriophages or phages, in the enological ecosystem (de Melo et al., 2018). However, grapes, and more globally the whole fermentation process, are an interesting crossroad between different ecosystems (insect, plant and its rhizosphere, soil and human), and may turn out to represent a valuable source for genome innovation for phages and their bacterial hosts. From the technological point of view, the characterization of phages from the enological niche could also represent an eco-friendly alternative to chemicals to limit bacterial spoilage during wine-making. Among established wine spoilers, acetic acid bacteria (AAB) negatively affect wine quality because of their ability to increase the volatile acidity of wine by the production of acetic acid among other compounds (Du Toit and Pretorius, 2000; De Roos and De Vuyst, 2018). Here, we detail a simple method for the screening and isolation of phages infecting AAB, that can be applied to other fruits (Philippe et al., 2018). Phage particles from small-scale production were concentrated by centrifugation, and time-consuming steps involving PEG precipitation or CsCl gradients were avoided. Cryo electron microscopy (cryo-EM) (Cuervo and Carrascosa, 2018) provided high resolution and distinctive features in the ultratructure of tectiviruses were visible.
Materials and Reagents
Materials
Autoclavable Erlenmeyer flasks (volume: 250 ml) (Dutscher, catalog number: 211907 )
Autoclavable glass tubes (volume: 20 ml) (Dutscher, catalog number: 0 45209 )
Cap-o-test caps (Dutscher, catalog number: 110685B )
Centrifuge tubes (volume: 50 ml) (Fisher Scientific, catalog number: 07-000-983 )
Pasteur pipette
Cryo Grid Box (Electron Microscopy Science, catalog number: 71166-10-W )
Freezer bags (450 ml, VWR, catalog number: 82007-706 )
Lacey Carbon 300 mesh copper grids (Ted Pella, catalog number: 01883)
Polyethersulfone (PES) membrane syringe filters (0.22 μm and 0.45 μm) (Fisher Scientific, catalog numbers: SLGPR33RS ; SLHPR33RS )
Polypropylene microcentrifuge tubes (volume: 1.5 ml) (Fisher Scientific, catalog number: 07-000-244 )
Spatula (Dutscher, catalog number: 442209 )
Spectrophotometer cuvettes (volume: 1.6 ml) (Dutscher, catalog number: 613101 )
Sterile disposable plastic syringes (volume: 50 ml) (Fisher Scientific, catalog number: 13-689-8 )
Sterile 90 x 13 mm Petri dishes (Dutscher, catalog number: 076084B )
Whatmann Filter paper grade 5 (Dutscher, catalog number: 1005055 )
Reagents
Agar-agar (Thermo Fisher Scientific, Fisher ChemicalTM, catalog number: 10548030 )
Calcium chloride dihydrate (CaCl2·2H2O) (Thermo Fisher Scientific, Fisher BioReagents, catalog number: 10306313 )
Ethane (AIR liquide)
Hydrochloric acid, 33.33% (w/v) aqueous solution (Thermo Fisher Scientific, ULTREX, J.T. BakerTM, catalog number: 10782232 )
Liquid nitrogen (cryo-distribution)
Magnesium sulfate heptahydrate (MgSO4·7H2O) (Thermo Fisher Scientific, Fisher BioReagents, catalog number: 10553335 )
Mannitol (Thermo Fisher Scientific, ACROS OrganicsTM, catalog number: 10266523 )
Peptone (Thermo Fisher Scientific, GibcoTM BactoTM Peptone, catalog number: 16299741 )
Sodium chloride (NaCl) (Thermo Fisher Scientific, Technical, catalog number: 10498220 )
Tris-HCl pH 7.5 (Thermo Fisher Scientific, Merck ChemicalsTM, catalog number: 15153685 )
Yeast extracts (Thermo Fisher Scientific, GibcoTM DifcoTM BactoTM, catalog number: 11763553 )
10x Phage buffer (see Recipes)
CaCl2·2H2O and MgSO4·7H2O 100x stock solutions (see Recipes)
YPM broth medium (see Recipes)
YPMΦ solid (“bottom”) medium (see Recipes)
YPMΦ soft agar (see Recipes)
Bacterial strain and samples of enological origin
Fresh crushed grapes (see Procedure)
Gluconobacter cerinus CRBO11179 (Centre de Resources Biologiques OEnologie, Institut des Sciences de la Vigne et du Vin, ISVV, Villenave-d’Ornon, France)
Equipment
Autoclave
Cryo-electron microscope (Thermo Fisher FEI, model: Tecnai F20 200 kV ) equipped with a CCD camera (Thermo Fisher FEI, model: Eagle 4kx4k)
Cryo-holder and cryo-transfer station (Gatan, model: 626 )
Glow Discharge unit (Cordouan Technologies, model: ELMO )
Incubator set to 25 °C
High-speed refrigerated centrifuge (Hitachi, model: CR 22N ) and R15A-0688 rotor
Microwave (Thermo Fisher Scientific, ManutanTM MW20G, catalog number: 15540739 )
Orbital shakers (Stuart, model: SSL1 )
pH meter (Hanna, model: pH211 )
Spectrophotometer (Shimadzu, model: UV-1280 )
Vitrification Robot (Leica Microsystem, model: EM-GP 2 )
Water bath (colume: 4 L) (Benchmark Scientific, MyBath model)
Software
ImageJ (NIH)
Tecnai version 4.3 (ThermoFisher FEI)
TEM Imaging & Analysis version 4.4 (ThermoFisher FEI)
Procedure
Collect grapes and recover juices
Collect fresh grapes into freezer bags (~200 x g) from selected vineyards/wineries.
Store at 4 °C during transport and process samples as quickly as possible.
Crush grapes manually and centrifuge the samples (5,000 x g, 10 min).
Collect the supernants (juices).
Filter the juices using successively 0.45 µm then 0.22 µm PES membrane filters under sterile conditions.
Store filtered juices in a 4 °C fridge.
Isolation of viral plaques, purification and production of a primary stock (Stock I)
Gluconobacter cerinus culture preparation
Prepare a sterile 250 ml Erlenmeyer flask with 15 ml of YPM broth.
Inoculate with 10 µl of a ready-to-use frozen bacterial strain in glycerol stock and incubate over night with shaking (200 rpm) at 25°C.
The following morning, back dilute the culture 1/10 into 15 ml of fresh YPMΦ broth and incubate at 25 °C in a shaking incubator until OD600nm 0.2 is obtained (4 to 5 h).
Preparation of the bottom top YPMΦ plates
Melt carefully the bottom top YPMΦ agar by heating in a microwave oven.
Cool down to 55 °C in a water bath.
Pour plates, using ~20 ml melted medium (prepare 3 plates per juice sample to analyze).
Allow the agar based media to solidify for 1 h at room temperature.
Double agar overlay assays
For each juice sample to analyze, melt 3 YPMΦ soft agar tubes in a boiling water bath.
Maintain the tubes in a 55 °C water bath prior to use.
Just before use, take the filtered juices out from the 4 °C fridge (Step A6) and prepare serial dilutions in phage buffer 1x (10-1, 10-2).
Remove a YPMΦ soft agar tube from the water bath and place it on a rack for 1-2 min at room temperature (RT).
Add 200 μl of bacterial culture at OD600nm 0.2 (Step B1) and 100 μl of undiluted filtered juice in the YPMΦ soft agar tube.
Vortex 3 times and spread evenly on top of a solid YPMΦ plate.
Repeat Steps B3d-f with each dilution (10-1, 10-2) of the juice samples (Step B3c) using the two remaining YPMΦ agar plates.
Incubate the plates at 25 °C for 24 h.
Isolation of plaques
Select the plate with individual plaques.
Visually identify the different morphotypes of plaques (large, small, clear, turbid) on the YPMΦ plate (if relevant).
Use a marker to draw a circle on the bottom of the plate around each selected plaque.
Punch each plaque out using a sterile Pasteur pipette with an aspiration bulb.
Place each recovered agar plug containing a single plaque into a micro-tube containing 500 μl of YPMΦ broth.
Vortex gently.
Allow the phages to diffuse in the broth medium for 20 min at RT.
Write R1 (first round of purification) on this microtube with a permanent marker.
Store at 4 °C or proceed immediately to Step B5.
Perform two additional rounds of purification
Dilute the R1 sample in phage buffer 1x (10-1 to 10-4) in micro-tubes.
Perform double layer agar plating for the 4 diluted samples (repeat Steps B1 to B3 for each diluted sample).
After incubation for 24 h, observe and select the plate which contains individual plaques.
Punch a single plaque into a micro-tube containing 500 µl of YPMΦ.
Write R2 (second round of purification) on your microtube with a permanent marker.
Repeat a third and last round of phage infection on the same sensitive bacterial strain to ensure purity. All steps need to be carried out under sterile conditions.
The 500 µl R3 sample is your primary pure phage stock (Stock I). Store it at 4 °C.
Preparation of a secondary phage stock (stock II) by using the confluent plate lysate method
This step is necessary to obtain an appropriate volume of phage lysate.
Double agar overlay assays
Prepare a 15 ml culture of G. cerinus CRBO 11179 culture (OD600nm 0.2) in YPMΦ broth (see Step B1).
Fill 3 sterile 1.5 ml microtubes with 900 µl of 1x sterile phage buffer and label the tubes with the dilution numbers (10-1 to 10-3).
Add 100 µl of stock I in the 10-1 tube and perform serial dilutions.
Prepare 24 YPMΦ agar plates.
Melt 24 YPMΦ soft agar tubes and place them in a water bath at 55 °C.
Remove 8 YPMΦ soft agar tubes from the water bath and place them on a rack for 1-2 min at room temperature (RT).
In each tube, mix 100 µl of the ten-fold phage stock I with 200 µl of the sensitive bacterium as shown in Steps B1 to B3.
Pour each tube on an YPMΦ agar plate.
Repeat Steps C1f to C1h with 8 additional soft agar tubes and YPMΦ agar plates and use 100 µl of stock I diluted to 10-2.
Repeat Steps C1f to C1h with the remaining 8 soft agar tubes and YPMΦ agar plates and use 100 µl of stock I diluted to 10-3.
Incubate the 24 plates 24 h at 25 °C.
Recovery of stock II
For each dilution plated on YPMΦ plates (10-1, 10-2, 10-3), observe the density of plaques.
Select the highest dilution giving confluent lysis (see Figure 1). Only the 8 plates corresponding to this dilution will be processed.
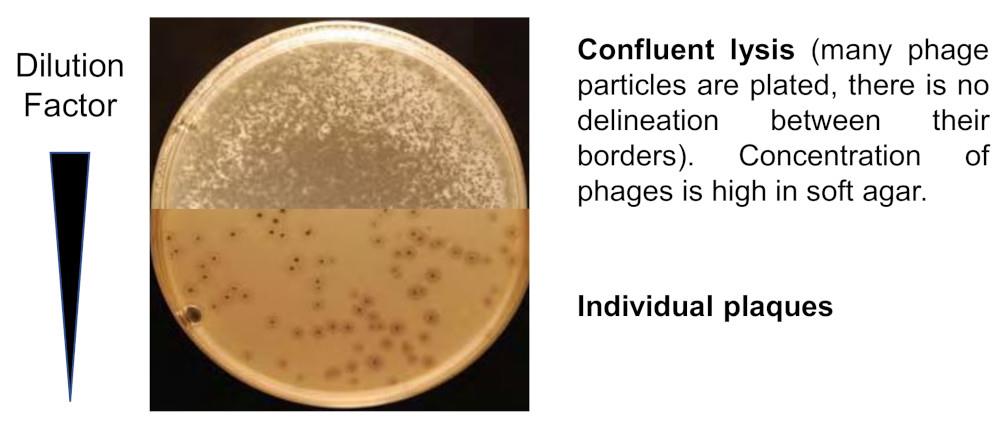
Figure 1. Ilustration of a confluent lysis
Under sterile conditions, add 1 ml of sterile YPMΦ broth on the surface of each of the 8 double layer agar plate selected.
Incubate 20 min at RT. Alternately, plates can be stored overnight at 4 °C at this step.
Use a sterile spatula to carefully collect the soft agars from each plate and the liquid medium in a centrifugation tube.
Centrifuge the sample at 10,000 x g for 10 min at 15 °C.
Filter the supernatant (6-8 ml) using 0.45 µm PES membrane filters.
Store stock II at 4 °C.
Determine the titer of stock II as follows (Figure 2)
Prepare a 15 ml culture of G. cerinus CRBO 11179 culture (OD600nm 0.2) in YPMΦ broth (see B1).
Prepare 8 sterile 1.5 ml micro-tubes containing 900 µl of 1x sterile phage buffe and serially dilute stock II 10-1 through 10-8.
Take an YPMΦ agar plate and draw a grid of 8-10 spaces on the plate bottom.
Remove a 5 ml YPMΦ soft agar tube from the water bath at 55 °C and inoculate with 0.2 ml of the culture prepared in Step C3a.
Vortex briefly to homogenize, pour on the YPMΦ agar plate and let the agar solidify at RT (30 min).
Spot a 10 µl drop of each dilution (pure to 10-8) of stock II in the available spaces of the grid; put the lid and let the Petri dish on the bench until the drops are adsorbed (1 h).
Incubate overnight at 25 °C.
Determine the dilution giving an optimal range to count between 2 to 20 plaques per space. The titer (PFU/ml) may be calculated by the following formula: PFU/ml (of secondary stock) = 1/dilution factor x number of plaques x 1/(0.01).

Figure 2. Rapid titration of phage stocks (II and III)
Preparation of highly concentrated GC1 phage suspension (stock III) by the liquid method
Introduce aseptically 15 ml YPMΦ broth in a sterile 250 ml Erlenmeyer flask.
Inoculate 10 µl of a frozen culture of G. cerinus CRBO11179.
Incubate overnight under 25 °C in a shaking incubator at 200 rpm.
Measure the OD600nm of the culture and transfer a volume in new flask containing 60 ml YPMΦ broth to obtain an OD600nm of 0.1, corresponding to approximately 108 CFU/ml.
Divide into three separate sterile 250 ml erlenmeyer flasks (20 ml/flask) (see Figure 3).

Figure 3. Summary of the preparation of phage stocks I, II and III
Infect two flasks with phage GC1 with a multiplicity of infection (MOI) of approximately 1 phage for 200 bacteria. The third culture is used as a non-infected negative control.
Allow the phages to adsorb to bacterial cells 30 min at RT without skaking.
Incubate at 25 °C in a shaking incubator at 200 rpm.
Measure the OD600nm periodically (mean values of 1 and 2.5 are observed for the infected assays and the control, respectively, after 14-16 h).
Note: Don’t extend incubation beyond 14-16 h as a 2-Log reduction in final phage titer is usually observed.
Centrifuge the lysed cultures at 10,000 x g for 10 min at 15 °C.
Filter the supernatant through a 0.22 μm PES filter.
Measure the titer of the lysate as described in Step C3.
Note: A titer of ~1010 PFU/ml is routinely obtained in the laboratory. The phage titer should exceed 107 PFU/ml to be observed under cryo-EM.
Transfer the filtered supernatant in a 50 ml centrifuge tube (ca. ~38 ml) and centrifuge 2 h at 20,000 x g at 4 °C (ensure all sample tubes are evenly filled to properly balance the centrifuge).
Carefully remove the supernatant and air dry the pellet.
Resuspend gently the phage pellet in 20 to 50 µl of sterile phage buffer 1x
Store at 4 °C.
Note: Sample should not be opaque for cryo-EM observation, if needed increase the dilution with phage buffer 1x.
Observation of GC1 phage particles by cryo-EM
We used the following standard protocol.
Vitrification: freezing of the preparation was made using the EM-GP apparatus (the procedure has been summarized Figure 4).
Fill the humidifier with distilled water and change the blotting paper using a Whatman grade 5.
Set the chamber conditions: temperature of 25 °C and 80% humidity and ethane temperature at -184 °C.
Set the forceps conditions: “blotter settings” must be adjusted to see the filter paper touching the grid without pulling it; “grid blot position” must be adjusted to place the grid 1 mm above the edge of the paper and “Grid Tf position” is adjusted to totally immersed the grid into the ethane container.
Adjust your freezing’s settings: Unselect the rotations and sensor. Select “A-plunge” to automatically plunge the grid after blotting. Set the time(s) settings as follows: Delay at 0.0; Blot at 2.0 s and Hold at 0.0.
Fill the container with liquid nitrogen until 100% after placing the ethane container and the grid box support.
Around -120 °C, condense the ethane at a pressure of 0.1 mBar.
Glow discharge the grids. Place the Lacey grids, carbon facing upwards, on a piece of glass slide covered with a Parafilm. Use the following glow discharge condition: vacuum at 0.3 mBar, intensity of 3 mA and timer of 40 s.
Apply 4 µl of the sample on the carbon side of the grid and activate the blotting and the plunge freezing by pressing “blot/A plunge”. Blotting is done on the opposite side of the sample drop.
Transfer the grid in the cryo-grid box. When all frozen grids are inside the grid box, screw the lid and transfer the grid box to a pre-cooled pierced 50 ml Falcon Tube under liquid nitrogen. Store the grid box container in a Dewar with liquid nitrogen until observation.
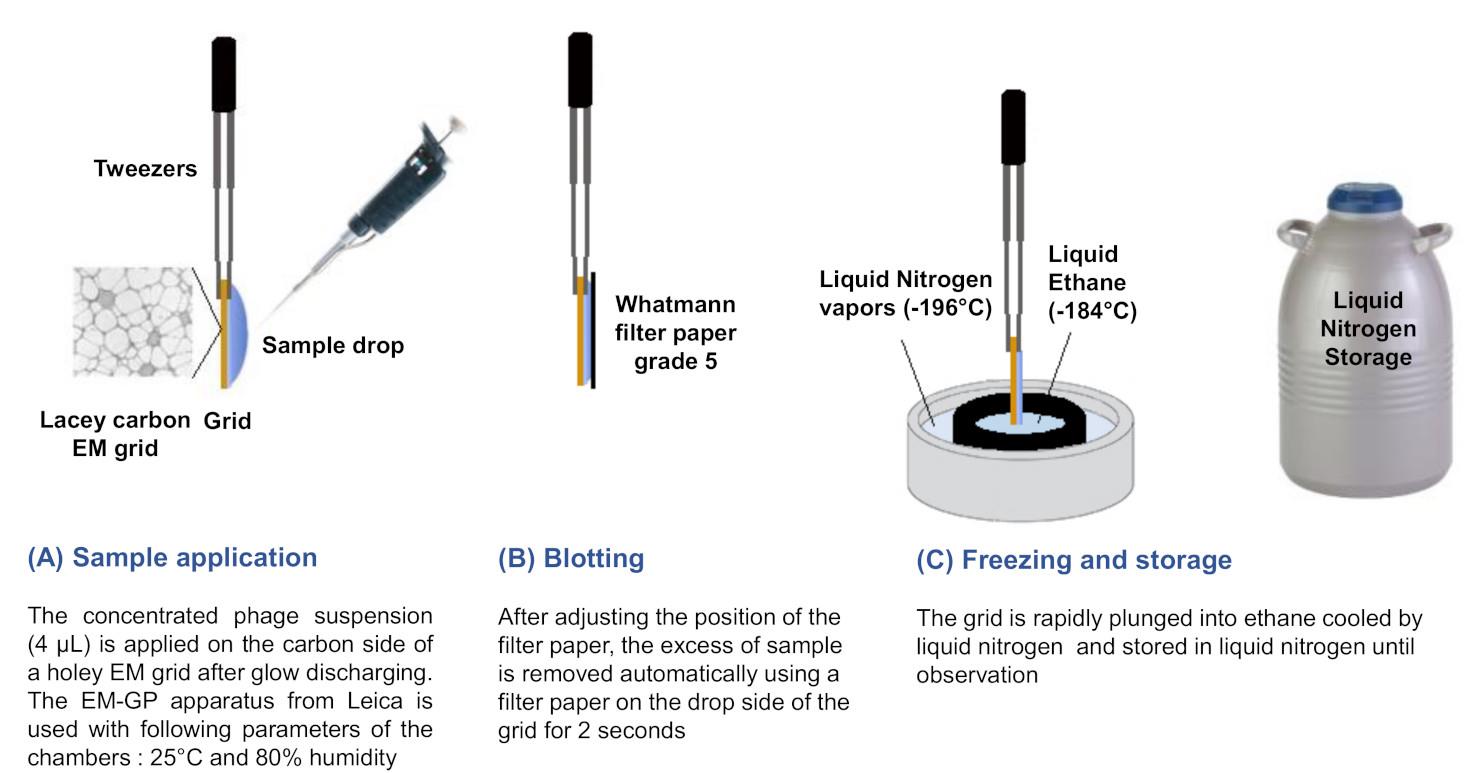
Figure 4. Graphical summary of the vitrification step
Cryo-EM observation: acquisitions were made using a Tecnai F20 operating at 200 kV (ThermoFisher FEI) equipped with a CCD camera (Eagle 4kx4k, ThermoFisher FEI)
Pump the GATAN 655 cryo-holder until the vacuum is better than 10-4 Torr.
Mount the grid on the cooled Cryo-holder.
Rotate the goniometer of the microscope to -55° to insert horizontally the holder to prevent all the liquid nitrogen going out. Carefully introduce the cryo-holder in the microscope and wait until the end of the count-down in the “vacuum overview”. Once the count-down in the “vacuum overview” has ended, introduce the cryo-holder completely into the microscope and rotate the goniometer to 0°.
Quickly refill the Dewar of the cryo-holder with liquid nitrogen.
Connect the cable of the control station to the cryo-holder and check the temperature around -175 °C.
Record images in low dose conditions and spot size 5. Search mode at x5,000 and exposure mode at x50,000 and 20 e-/Å2s.
Data analysis
Reliable measurements of the phages morphology and sizes were obtained from cryo-TEM images and performed with ImageJ software.
Data analyses
Plunge freezing of GC1 particles allows us to observe their exact shape with reduced shrinkage of the capsid (Figures 5A-5D). GC1 is a tailless phage of about 60 nm in diameter, with the presence of an inner lipid membrane (Figure 5D, white arrow). The virion presents eleven vertices on its surface where spike complexes are anchored. A unique vertex ensures the viral DNA packaging and injection through the formation of a tubular structure from the membrane of about 10 nm in diameter and 60 nm in length (Figure 5D, black arrow). When DNA injection is not triggered, viral particles are fully filled with genetic material (Figure 5C) while it is partially filed when the tubular structure occurs (Figure 5D) (Saren et al., 2005; Peralta et al., 2013). We refer the readers to the original paper (Philippe et al., 2018).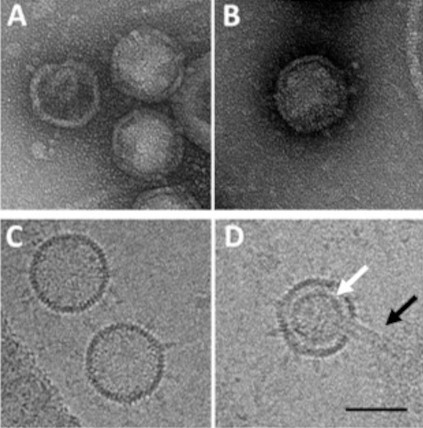
Figure 5. Comparative images of negatively stained and cryo-fixed GC1 phage particles. A-B: GC1 phage prepared by negative staining and observed by TEM (Philippe et al., 2018). C-D: GC1 phage prepared by plunge freezing and observed by cryo-EM. Scale bar = 50 nm.
Notes
Acetic acid bacteria (AAB) are generally recognized as safe bacteria and all the experiments can be performed in a biosafety level 1 laboratory. AAB can be easily recovered from mature/overippe grapes, and isolated on YPM agar supplemented with 0.1 ml of a 0.25% solution of penicillin (to inhibit the growth of Lactic acid bacteria) and 0.2 ml of a 0.25% alcoholic solution of pimaricin (to suppress the growth of yeasts and moulds). 5
Alternatively, musts can be collected from wineries (instead of grapes).
We analyzed several musts from red and white grapes. A single sample from white grapes yielded plaques on G. cerinus CRBO11179. All had the same morphotype. Two individual plaques were isolated and both corresponded to phage GC1. 5
During preparation of stock I (Steps B4 and B5), intermediate R1, R2 and R3 lysates can be stored at 4 °C for a few weeks.
Don’t use chloroform to avoid bacterial contamination of your phage stocks as it leads to Tectiviridae phage particles inactivation.
Phage titer of stock II is 1,000 to 10,000-fold higher than that of stock I at this step.
Step C produces a sufficient volume of fresh crude phage stock (stock II) to prepare a more concentrated lysate (stock III) by the Liquid Method (Procedure D). Stock II is not suitable for visual analysis by cryo EM.
Recipes
Phage Buffer 10x (for 1 L)
Dissolve 58 g of NaCl and 20 g of MgSO4·7H2O in 500 ml of 1 M Tris-HCl pH 7.5
Complete to 1 L with distilled water
Sterilize by autoclaving
Store at room temperature
To prepare 1x phage buffer, dilute 10 ml of 10x phage buffer with 90 mL distilled water and sterilize it by autoclaving (1 bar, 20 min)
CaCl2·2H2O and MgSO4·7H2O 100x stock solutions
Dissolve 35.5 g of CaCl2·2H2O in 100 ml of distilled water
Dissolve 19.3 g of MgSO4·7H2O in 100 ml of distilled water
Sterilize both stock solutions at 1 bar for 15 min
Yeast-Peptone-Mannitol (YPM) broth (for 1 L)
Weight 5 g of yeast extract, 3 g of peptone and 25 g of mannitol
Add 800 ml distilled water
Adjust pH to 5 with hydrochloric acid aqueous solution
Complete to 1 L with distilled water
Sterilize by autoclaving (1 bar, 20 min)
Modified Yeast-Peptone-Mannitol solid medium (YPMΦ “bottom” agar) (for 1 L)
Prepare 1 L of YPM broth (3a. to c.)
Add 10 ml of CaCl2·2H2O and MgSO4·7H2O 100x stock solutions
Add 20 g of granulated agar
Complete to 1 L with distilled water
Sterilize by autoclaving (1 bar, 20 min)
Store at room temperature
Modified Yeast-Peptone-Mannitol soft agar medium (YPMΦ) (for 1 L)
Prepare 1 L of YPMΦ broth (4a. to c.)
Add 6 g of granulated agar
Dissolve agar by heating in a microwave until complete melting of the agar
Distribute in 5 ml aliquots in glass tubes (20 ml) and close with cap-o-test caps
Sterilize by autoclaving
Store at 4 °C
References
- Cuervo, A. and Carrascosa, J. L. (2018). Observation of bacteriophage ultrastructure by cryo-electron microscopy. Methods Mol Biol 1693: 43-55.
- de Melo, A. G., Levesque, S. and Moineau, S. (2018). Phages as friends and enemies in food processing. Curr Opinion Biotechnol 49: 185-190.
- De Roos, J. and De Vuyst, L. (2018). Acetic acid bacteria in fermented foods and beverages. Curr Opinion Biotechnol 49: 115-119.
- Du Toit, M. and Pretorius, I. (2000). Microbial spoilage and preservation of wine: Using weapons from Nature's own arsenal-A review. S Afr J Enol Vitic 21: 74-96.
- Peralta, B., Gil-Carton, D., Castaño-Díez, D., Bertin, A., Boulogne, C., Oksanen, H. M., Bamford, D. H. and Abrescia, N. G. (2013). Mechanism of membranous tunnelling nanotube formation in viral genome delivery. PLoS biology 11(9): e1001667.
- Philippe, C., Krupovic, M., Jaomanjaka, F., Claisse, O., Petrel, M. and le Marrec, C. (2018). Bacteriophage GC1, a Novel Tectivirus Infecting Gluconobacter cerinus, an Acetic Acid Bacterium Associated with Wine-Making. Viruses 10(1).
- Saren, A. M., Ravantti, J. J., Benson, S. D., Burnett, R. M., Paulin, L., Bamford, D. H. and Bamford, J. K. (2005). A snapshot of viral evolution from genome analysis of the tectiviridae family. J Mol Biol 350(3): 427-440.
Article Information
Copyright
© 2020 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Chaïb, A., Decossas, M., Philippe, C., Claisse, O., Lambert, O. and Le Marrec, C. (2020). Isolation and CryoTEM of Phages Infecting Bacterial Wine Spoilers. Bio-protocol 10(21): e3801. DOI: 10.21769/BioProtoc.3801.
Category
Microbiology > Microbe-host interactions > Bacterium
Cell Biology > Cell isolation and culture > Virus isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link