- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Biofilm Formation Assay in Pseudomonas syringae
Published: Vol 9, Iss 10, May 20, 2019 DOI: 10.21769/BioProtoc.3237 Views: 13736
Reviewed by: Anastasia D GaziAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Purification of the Bacterial Amyloid “Curli” from Salmonella enterica Serovar Typhimurium and Detection of Curli from Infected Host Tissues
Murugesan Sivaranjani [...] Aaron P. White
May 20, 2022 3233 Views

A Guideline for Assessment and Characterization of Bacterial Biofilm Formation in the Presence of Inhibitory Compounds
Bassam A. Elgamoudi and Victoria Korolik
Nov 5, 2023 3104 Views

Surface Plasmon Resonance for the Interaction of Capsular Polysaccharide (CPS) With KpACE
Zhe Wang [...] Chao Cai
Jun 20, 2025 3552 Views
Abstract
Pseudomonas syringae is a model plant pathogen that infects more than 50 plant species worldwide, thus leading to significant yield loss. Pseudomonas biofilm always adheres to the surfaces of medical devices or host cells, thereby contributing to infection. Biofilm formation can be visualized on numerous matrixes, including coverslips, silicone tubes, polypropylene and polystyrene. Confocal laser scanning microscopy can be used to visualize and analyze biofilm structure. In this study, we modified and applied the current method of P. aeruginosa biofilm measurement to P. syringae, and developed a convenient protocol to visualize P. syringae biofilm formation using a borosilicate glass tube as the matrix coupled with crystal violet staining.
Keywords: BiofilmBackground
Most Pseudomonas strains secrete exopolysaccharides, such as alginate, which is an important matrix molecule for biofilm formation (Hentzer et al., 2001; Nivens et al., 2001). Biofilm formed by the human pathogen P. aeruginosa plays important roles in its virulence and antibiotic resistance, and contributes to acute or chronic infections (Donlan and Costerton, 2002).
To date, various methods have been reported for biofilm characterization and quantification. Originally, biofilms were detected in microtiter plates made of polystyrene or polypropylene (O'Toole and Kolter, 1998; Merritt et al., 2005). During the growth of P. aeruginosa on a surface, the expression of genes involved in extracellular polysaccharide synthesis is induced (Davies et al., 1993; Davies et al., 1995), which promotes the adherence of cells to the surface. Crystal violet specifically stains the bacterial cells, and has been developed as a widely used dye for bacterial biofilm (George et al., 1998). Some recent studies have analyzed biofilm structure in a flow chamber coupled with confocal laser scanning microscopy (Sternberg and Tolker-Nielsen, 2006; Chua et al., 2016).
Biofilms formed by P. syringae strains have also been found in plant tissues (Osman et al., 1986; Fakhr et al., 1999; Preston et al., 2001). Alginate produced by P. syringae is an important polymer for P. syringae biofilm formation and contributes to its virulence and fitness, indicating its importance in plant-pathogen interaction (Preston et al., 2001; Engl et al., 2014). The formation of P. syringae and P. fluorescens biofilms can also be measured using crystal violet staining in microwell plates (Carezzano et al., 2017; Zhu et al., 2018; Patange et al., 2019).
In this study, we modified and applied the current method of P. aeruginosa biofilm measurement to P. syringae (Kong et al., 2015; Zhao et al., 2016; Shao et al., 2018). We present an economic, rapid and visual biofilm detection protocol that combines the use of borosilicate glass tubes and crystal violet staining methods, which have been efficiently used in our recent studies (Wang et al., 2018; Wang et al., 2019; Xie et al., 2019) for visualizing the biofilm of the model plant pathogen P. syringae.
Materials and Reagents
- 10 ml Borosilicate glass tube (ISOLAB, catalog number: 077.02.003)
- 14 ml sterile tube (SPL Lifescience, catalog number: 40014)
- Filter (PALL Lifesciences, catalog number: AP-4219)
- Strains P. syringae pv. phaseolicola 1448A (Psph) (Xiao et al., 2007) and rhpS deletion mutant (ΔrhpS) (Xie et al., 2019)
- NaOH (UNI-CHEM, catalog number: 1310-73-2)
- MgSO4·7H2O (Aladdin, catalog number: 10025-84-0)
- K2HPO4 (Aladdin, catalog number: 7758-11-4)
- BactoTM Proteose peptone No.3 (AOBOX, catalog number: 01-049)
- Rifampin (Aladdin, catalog number: 13292-46-1)
- Agar (MP Biomedicals, catalog number: 9002-18-0)
- Crystal violet (Beijing Dingguo, catalog number: 548-62-9)
- Glycerol (Beijing Bailingwei, catalog number: 262536)
- 100% ethanol (Honeywell, catalog number: 32221-2.5L)
- King's B (KB) (see Recipes)
Equipment
- 1 ml pipette (Eppendorf, catalog number: 3123000063)
- Benchtop shaking incubator (Labwit Scientific, model: ZWYR-240)
- Constant temperature incubator (Labwit Scientific, model: ZXDP-B2120)
- EvolutionTM 350 UV-Vis Spectrophotometer (Thermo Fisher Scientific, catalog number: 912A0959)
- SynergyTM 2 Multi-Mode Microplate Reader (BioTek)
- Test tube stand (ISOLAB, catalog number: 079.01.005)
- -80 °C freezer (Thermo Scientific, catalog number: 5IDTSX)
Software
- Microsoft Office Excel 2016 and GraphPad Prism 8.0.2.
Procedure
Notes:
- Select wild-type P. syringae pv. phaseolicola 1448A (Psph) as the model strain (Xiao et al., 2007) and rhpS deletion mutant (ΔrhpS) (Xie et al., 2019) as the test strain.
- Perform the step-by-step protocol described in Figure 1.
- Perform the entire procedure gently to avoid damaging the biofilm.
- Collect the liquid crystal violet and ethanol waste liquor in specially labeled containers for professional disposal by trained staff, as per the safety regulations at City University of Hong Kong.

Figure 1. Schematic step-by-step protocol for visualizing P. syringae biofilm formation. A. The Psph strain was activated on King’s B (KB) plate and cultured in liquid medium. B. Then the cultures were inoculated at 1:500 dilutions into KB liquid medium and incubated statically to sampling points. C. Then stained the biofilm by using 0.1% crystal violet. D. Measure the biofilm production at OD590nm. Two biological replicates were showed.
- Bacterial growth
- Collect a Psph colony from the glycerol stock culture frozen at -80 °C and inoculate on a King’s B (KB) plate supplemented with rifampicin (25 μg/ml). Incubate the KB plate at 28 °C for 36 h in a constant temperature incubator.
- Collect a single colony from the cultured plates and inoculate into a sterile 10 ml tube containing 2 ml KB liquid medium supplemented with rifampicin (25 μg/ml). Incubate the tube in a benchtop shaking incubator at 28 °C for 12 h with constant shaking at 220 rpm.
- Biofilm formation
Inoculate the Psph culture (1:500 dilutions) into 16 sterile 10 ml borosilicate glass tubes containing 2 ml KB liquid medium (supplemented with rifampicin) and ensure consistent initial dose. Incubate the Psph-containing glass tubes at 28 °C in a constant temperature incubator without shaking. - Biofilm visualization
- Harvest the biofilm samples at different time points. For Psph in our study, the biofilms were harvested at 24, 48, 72 and 96 h. Gently discard the planktonic cells with a 1 ml pipette and wash the tubes three times with sterile distilled water. Avoid damaging the biofilm formed on the tube wall.
- Stain the biofilm forming bacteria with 2.5 ml 0.1% crystal violet for 20 min without shaking. Discard the dye and wash the tubes with sterile distilled water to remove the unbound dye. Dry the tubes and take photographs (Figure 2A).
- Biofilm measurement
- Elute the biofilm with 2 ml 100% ethanol and shake the tubes at 220 rpm for 20 min to ensure that the dye has dissolved completely. Take photographs (Figure 2B).
- Measure the eluted samples at OD590nm using a spectrophotometer (2 ml) or Synergy 2 Plate Reader (BioTek) (100 μl). If the sample concentrations are too high, diluted them before measuring. Use an equal volume of 95%-100% ethanol as the blank control. Psph biofilm formation is shown in Figure 2C. The ΔrhpS produces lower biofilm compared with Psph wild-type strain (Figure 3).

Figure 2. Visualization and quantification of biofilm in Psphwild-type strain. A. Biofilm samples were grown from 24 to 96 h. Biofilm adhered to borosilicate glass tubes at different time points were stained with crystal violet. B. The crystal violet bound to the biofilm on the wall of the tubes was eluted by ethanol. C. The elution samples were measured at OD590nm by using spectrophotometer or Synergy 2 Plate Reader (BioTek). Psph wild-type strain produced more biofilm at 96 h than 24 h. * represents P-value < 0.05. *** represents P-value < 0.001. Error bars indicate S.D. among four biological replicates. Two biological replicates were shown.
Figure 3. The ΔrhpS strain produced less biofilm than did that in Psph wild-type strain. A. Biofilm produced by the Psph wild-type and the ΔrhpS strain were visualized using borosilicate glass tubes and stained with crystal violet at 96 h. B. The crystal violet bound to the biofilm on tube wall was eluted by ethanol. C. The Psph wild-type strain produced more biofilm than the ΔrhpS strain (P-value = 0.000136). *** represents P-value < 0.001. Error bars indicate S.D. among four biological replicates. Two biological replicates were shown.
Data analysis
Student’s t-tests were performed using Microsoft Office Excel 2016. Quantitative data (OD590nm) were collected from four biological replicates in the figures and tables (Tables 1 and 2).
Table 1. Biofilm production of Psph wild-type strain presented in Figure 2C. Two-sample equal variance was calculated by the following one-tailed Student’s t-test formula in Excel = TTEST (array1, array2, tails, type). Array 1 is the first data set. Array 2 is the second data set. Tails show the number of distribution tails (1 for the one-tailed distribution, 2 for two-tailed distribution). Type is the kind of t-test to perform (1 for paired, 2 for two-sample equal variance, and 3 for two-sample unequal variance. For example, we used TTEST (B2:E2, B3:E3, 1, 2), TTEST (B2:E2, B4:E4, 1, 2) and TTEST (B2:E2, B5:E5, 1, 2) for biofilm production at 48 h, 72 h, 96 h, compared to 24 h respectively. The P-values were showed in Excel H3, H4 and H5 respectively. * represents P-value < 0.05. ** represents P-value < 0.01. *** represents P-value < 0.001. Means and S.D. are shown in columns F and G. All experiments were repeated four times.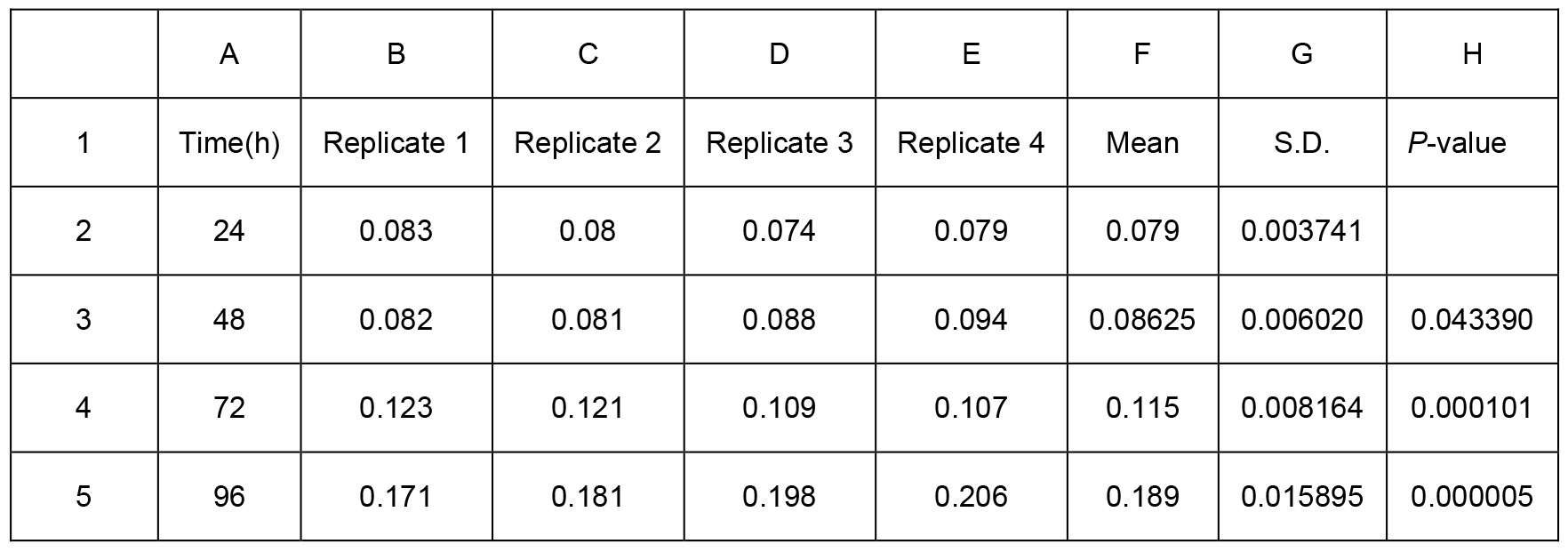
Table 2. Biofilm production of the Psph wild-type and the ΔrhpS strain presented in Figure 3C. Two-sample equal variance was calculated by the following Two-tailed Student’s t-test formula in Excel = TTEST (B2:E2, B3:E3, 2, 2). The P-values were showed in Column H3. * represents P-value < 0.05. ** represents P-value < 0.01. *** represents P-value <0.001. Means and S.D. are shown in Column F and G. All experiments were repeated four times.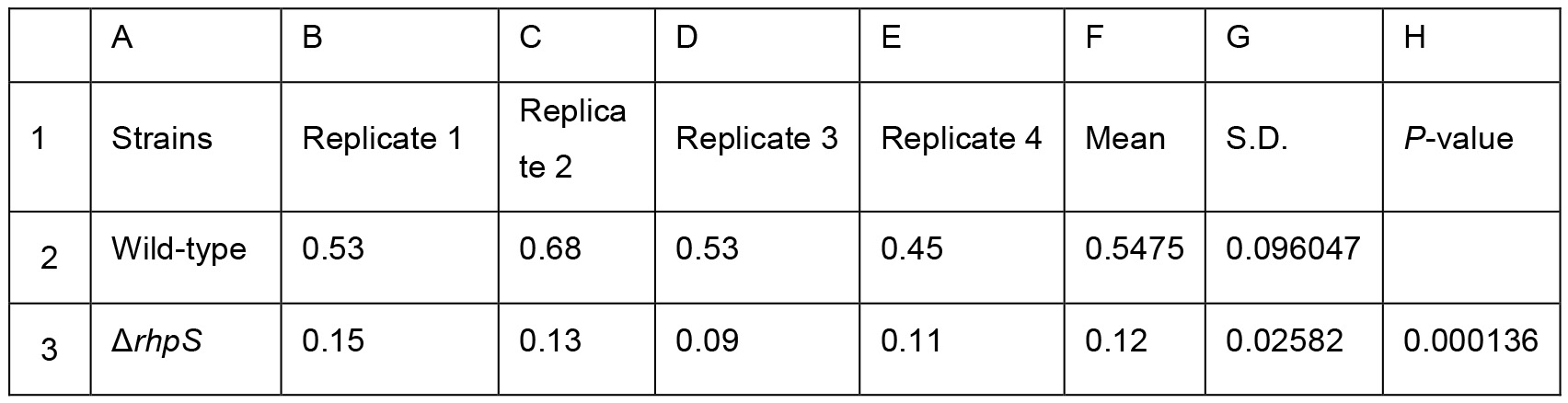
In sum, the results showed that the wild-type Psph strain produced more biofilm in 48 h (P < 0.05), 72 h (P < 0.001) and 96 h (P < 0.001) than at 24 h (Figure 2 and Table 1). Besides, the ΔrhpS strain produced less biofilm than Psph wild-type strain (P < 0.001) at the same time point (96 h) (Figure 3 and Table 2).
Recipes
- King's B (KB) (King et al., 1954)
Dissolve in 993.75 ml ddH2O and adjust the pH to 7.2. Add 15 g/L agar for solidified media and autoclaveBactoTM Proteose peptone No.3 20.0 g/L K2HPO4 1.5 g/L Glycerol 15 ml/L
Dissolve 24.637 g MgSO4·7H2O in 100 ml ddH2O and sterilize this stock solution (1 M) using sterile filter (0.45 µm)
Then, add 6.25 ml MgSO4·7H2O (1 M) (final concentration 1.5 g/L) into the autoclaved King's B medium when the temperature drops to 40 °C-50 °C
Acknowledgments
This project has been funded in Health Medical Research Fund [17160022 to X.D.]; National Natural Science Foundation of China Grants [31670127 to X.D.].
This protocol was adapted from O'Toole et al. (1998) Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Molecular Microbiology, 30: 295-304.
Competing interests
Conflict of interest statement: None declared.
References
- Carezzano, M. E., Sotelo, J. P., Primo, E., Reinoso, E. B., Paletti Rovey, M. F., Demo, M. S., Giordano, W. F. and Oliva, M. L. M. (2017). Inhibitory effect of Thymus vulgaris and Origanum vulgare essential oils on virulence factors of phytopathogenic Pseudomonas syringae strains. Plant Biol (Stuttg) 19(4): 599-607.
- Chua, S. L., Yam, J. K., Hao, P., Adav, S. S., Salido, M. M., Liu, Y., Givskov, M., Sze, S. K., Tolker-Nielsen, T. and Yang, L. (2016). Selective labelling and eradication of antibiotic-tolerant bacterial populations in Pseudomonas aeruginosa biofilms. Nat Commun 7: 10750.
- Donlan, R. M. and Costerton, J. W. (2002). Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15(2): 167-193.
- Davies, D. G., Chakrabarty, A. M. and Geesey, G. G. (1993). Exopolysaccharide production in biofilms: substratum activation of alginate gene expression by Pseudomonas aeruginosa. Appl Environ Microbiol 59(4): 1181-1186.
- Davies, D. G. and Geesey, G. G. (1995). Regulation of the alginate biosynthesis gene algC in Pseudomonas aeruginosa during biofilm development in continuous culture. Appl Environ Microbiol 61(3): 860-867.
- Engl, C., Waite, C. J., McKenna, J. F., Bennett, M. H., Hamann, T. and Buck, M. (2014). Chp8, a diguanylate cyclase from Pseudomonas syringae pv. Tomato DC3000, suppresses the pathogen-associated molecular pattern flagellin, increases extracellular polysaccharides, and promotes plant immune evasion. MBio 5(3): e01168-01114.
- Fakhr, M. K, Penaloza-Vazquez, A, Chakrabarty, A. M, Bender, C. L. (1999). Regulation of alginate biosynthesis in Pseudomonas syringae pv. syringae. J Bacteriol 181:3478-3485.
- Hentzer, M., Teitzel, G. M., Balzer, G. J., Heydorn, A., Molin, S., Givskov, M. and Parsek, M. R. (2001). Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J Bacteriol 183(18): 5395-5401.
- King, E. O., Ward, M. K. and Raney, D. E. (1954). Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med 44(2): 301-307.
- Kong, W., Zhao, J., Kang, H., Zhu, M., Zhou, T., Deng, X. and Liang, H. (2015). ChIP-seq reveals the global regulator AlgR mediating cyclic di-GMP synthesis in Pseudomonas aeruginosa. Nucleic Acids Res 43(17): 8268-8282.
- Merritt, J. H., Kadouri, D. E. and O'Toole, G. A. (2005). Growing and analyzing static biofilms. Curr Protoc Microbiol Chapter 1: Unit 1B 1.
- Nivens, D. E., Ohman, D. E., Williams, J. and Franklin, M. J. (2001). Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J Bacteriol 183(3): 1047-1057.
- Osman, S. F., Fett, W. F. and Fishman, M. L. (1986). Exopolysaccharides of the phytopathogen Pseudomonas syringae pv. glycinea. J Bacteriol 166(1): 66-71.
- O'Toole, G. A. and Kolter, R. (1998). Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30(2): 295-304.
- Patange, A., Boehm, D., Ziuzina, D., Cullen, P. J., Gilmore, B. and Bourke, P. (2019). High voltage atmospheric cold air plasma control of bacterial biofilms on fresh produce. Int J Food Microbiol 293: 137-145.
- Preston, L. A., Bender, C. L. and Schiller, N. L. (2001). Analysis and expression of algL, which encodes alginate lyase in Pseudomonas syringae pv. syringae. DNA Seq 12(5-6): 455-461.
- Shao, X., Zhang, X., Zhang, Y., Zhu, M., Yang, P., Yuan, J., Xie, Y., Zhou, T., Wang, W., Chen, S., Liang, H. and Deng, X. (2018). RpoN-dependent direct regulation of quorum sensing and the Type VI secretion system in Pseudomonas aeruginosa PAO1. J Bacteriol 200(16).
- Sternberg, C. and Tolker-Nielsen, T. (2006). Growing and analyzing biofilms in flow cells. Curr Protoc Microbiol Chapter 1: Unit 1B 2.
- Wang, J., Shao, X., Zhang, Y., Zhu, Y., Yang, P., Yuan, J., Wang, T., Yin, C., Wang, W., Chen, S., Liang, H. and Deng, X. (2018). HrpS is a global regulator on Type III Secretion System (T3SS) and Non-T3SS genes in Pseudomonas syringae pv. phaseolicola. Mol Plant Microbe Interact 31:1232-1243.
- Wang, T., Cai, Z., Shao, X., Zhang, W., Xie, Y., Zhang, Y., Hua, C., Schuster, S. C., Yang, L. and Deng, X. (2019). The pleiotropic effects of c-di-GMP content in Pseudomonas syringae. Appl Environ Microbiol pii: AEM.00152-19.
- Xiao, Y., Lan, L., Yin, C., Deng, X., Baker, D., Zhou, J. M. and Tang, X. (2007). Two-component sensor rhpS promotes induction of Pseudomonas syringae type III secretion system by repressing negative regulator RhpR. Mol Plant Microbe Interact 20(3): 223-234.
- Xie, Y., Shao, X., Zhang, Y., Liu, J., Wang, T., Zhang, W., Hua, C., Deng, X. (2019). Pseudomonas savastanoi two-component system RhpRS switches between virulence and metabolism by tuning phosphorylation state and sensing nutritional conditions. mBio 10(2). pii: e02838-18.
- Zhao, J., Yu, X., Zhu, M., Kang, H., Ma, J., Wu, M., Gan, J., Deng, X. and Liang, H. (2016). Structural and molecular mechanism of CdpR involved in Quorum-sensing and bacterial virulence in Pseudomonas aeruginosa. PLoS Biol 14(4): e1002449.
- Zhu, J., Yan, Y., Wang, Y. and Qu, D. (2019). Competitive interaction on dual-species biofilm formation by spoilage bacteria, Shewanella baltica and Pseudomonas fluorescens. J Appl Microbiol 126(4): 1175-1186.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Shao, X., Xie, Y., Zhang, Y. and Deng, X. (2019). Biofilm Formation Assay in Pseudomonas syringae. Bio-protocol 9(10): e3237. DOI: 10.21769/BioProtoc.3237.
Category
Microbiology > Microbial biofilm > Biofilm culture
Biochemistry > Carbohydrate > Polysaccharide
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










