- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
TZA, a Sensitive Reporter Cell-based Assay to Accurately and Rapidly Quantify Inducible, Replication-competent Latent HIV-1 from Resting CD4+ T Cells
Published: Vol 9, Iss 10, May 20, 2019 DOI: 10.21769/BioProtoc.3232 Views: 7794
Reviewed by: Emilie Besnard Mauricio MontanoAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In vitro Assays for Measuring Endothelial Permeability by Transwells and Electrical Impedance Systems
Hong-Ru Chen and Trai-Ming Yeh
May 5, 2017 28043 Views

Isolation, BODIPY Labeling and Uptake of Exosomes in Hepatic Stellate Cells
Shubham Shrivastava
Dec 5, 2017 12238 Views

Generation and Implementation of Reporter BHK-21 Cells for Live Imaging of Flavivirus Infection
Jorge L. Arias-Arias and Rodrigo Mora-Rodríguez
Mar 5, 2021 4367 Views
Abstract
The latent HIV-1 viral reservoir in resting CD4+ (rCD4+) T cells represents a major barrier to an HIV-1 cure. There is an ongoing effort to identify therapeutic approaches that will eliminate or reduce the size of this reservoir. However, clinical investigators lack an assay to determine whether or not a decrease in the latent reservoir has been achieved. Therefore, it is critical to develop assays that can reproducibly quantify the reservoir size and changes therein, in participant’s blood during a therapeutic trial. Quantification of the latent HIV viral reservoir requires a highly sensitive, cost-effective assay capable of measuring the low frequency of rCD4+ T cells carrying functional provirus. Preferably, such an assay should be such that it can be adopted for high throughput and could be adopted under conditions for use in large-scale clinical trials. While PCR-based assays are commonly used to quantify pro-viral DNA or intracellular RNA transcript, they cannot distinguish between replication-competent and defective proviruses. We have recently published a study where a reporter cell-based assay (termed TZA or TZM-bl based quantitative assay) was used to quantify inducible replication-competent latent HIV-1 in blood. This assay is more sensitive, cost-efficient, and faster than available technology, including the quantitative viral outgrowth assay or the Q-VOA. Using this assay, we show that the size of the inducible latent HIV-1 reservoir in virally suppressed participants on ART is approximately 70-fold larger than previous estimates. We describe here in detail an optimized method to quantitate latently infected cells using the TZA.
Keywords: TZM-bl cellsBackground
The ability to quantitate the latent HIV-1 viral reservoir in a combination ART suppressed individual requires a highly sensitive assay with the ability to measure low frequency of rCD4+ cells carrying functional provirus. This assay should also be cost-effective and adaptable to be high throughput under different conditions and large-scale clinical applications. Currently, the frequency of the infected cells in latent condition is estimated using Poisson Statistics or by maximum likelihood analysis (Cillo et al., 2014; Rosenbloom et al., 2015). Most of these methods including the original Q-VOA method and its adaptations use limited dilution based techniques of PHA simulated CD4+ T cells which either measures HIV-1 protein or RNA via ELISA or quantitative PCR. These are all labor-intensive, time-consuming, expensive, and requires the frequent addition of activated CD4+ T cells as feeders (Chun et al., 1997; Finzi et al., 1999; Siliciano et al., 2003; Siliciano and Siliciano, 2005). There has also been a modified version of Q-VOA assay reported which implements MOLT-4/CCR5 (Laird et al., 2013) cell line for viral expansion instead of activated CD4+ T cells followed by RNA measurement but this is still more time consuming and labor intensive.
We recently reported the development of a sensitive TZM-bl cell-based assay (termed TZA) (Sanyal et al., 2017) to quantify the latent HIV-1 reservoir in blood. The present protocol consists of these main parts (i) isolation of resting T cells from the blood from HIV-1 positive and negative (control) donors (Steps A1-A4); (ii) activation of these resting CD4+ T cells using a strong LRA like Anti-CD3/CD28 (Siliciano and Siliciano, 2004) antibodies and (iii) washing, counting and plating these activated cells on a TZM-bl reporter cells to quantitate replication-competent HIV-1 by measuring β-gal expression to quantify virus induced. This assay utilizes the TZM-bl cell line, which stably expresses CD4, CCR5, and CXCR4, and carries an integrated copy of the β-galactosidase (β-gal) gene under the control of an HIV-1 long terminal repeat (HIV-1 LTR) promoter that allows for the detection of inducible replication-competent HIV-1. By using TZM-bl cells, quantification of replication-competent HIV-1 can be achieved with high sensitivity (Ananworanich and Mellors, 2015). TZA has been used to calculate fraction of inducible latent virus (fPVE) (Cillo et al., 2014; Sanyal et al., 2017). The level of fPVE can be used to screen various LRAs in-vitro as well as help monitor their in-vivo efficacy especially during clinical trials. This assay reveals that the viral reservoir is likely much larger than previously predicted and estimated which can impact the ongoing therapeutic approaches to eradicate HIV-1.
This assay can be adapted and automated to a 384-well format further down the line to enable an efficient screening platform, which will be capable of handling multiple samples simultaneously. This will make it a high-throughput assay system. In addition, because of the low cell requirement in this system, the TZA may also be useful for quantification of replication-competent HIV-1 in the pediatric population as well as estimation of the reservoir in tissues. We are presently developing a protocol for determining the reservoir size in tissues using this assay.
Materials and Reagents
- Pipette tips
- 15 ml centrifuge tubes (Fisherbrand, catalog number: 07-200-886)
- 50 ml centrifuge tubes (Fisherbrand, catalog number: 0553913)
- Tissue culture plates, 24-well transparent (Falcon, catalog number: 353047)
- White, 96-well plates, sterile with white lids (PerkinElmer, catalog number: 6005181)
- EDTA tubes purple tops for blood collection: Vacutainer brand sterile (BD, catalog number: 366643)
- CryoTubeTM vials for freezing cells (Thermo Fischer Scientific, catalog numbers: 153779, 202209, 363401)
- Tissue culture plates, 48-well transparent (Falcon, catalog number: 353078)
- 5 ml round-bottom polystyrene tubes (Corning, Falcon®, catalog number: 352054)
- TZM-bl cells (NIH AIDS Reagent program, catalog number: 8129)
- Animal serum complex (Gemini Bio Products, Fetal PlexTM, catalog number: 100-602), storage in a -18 °C freezer before opening bottle and then store at 4 °C for subsequent use
- DMSO (Life Technologies Corp, catalog number: 20688)
- FBS (HyClone, catalog number: SH3011803)
- Beta-Glo Assay System (Promega, catalog number: E4780), storage in a -18 °C freezer
- Custom ordered resting CD4+ T cells negative selection kit (Stemcell Technologies, catalog number: 19309VK) with EasySep D Magnetic Particles (Stemcell Technologies, catalog number: 19250), storage at 4 °C
- DynabeadsTM Human T-Activator CD3/CD28 (Thermo Fischer Scientific, GibcoTM, catalog number: 11131D)
- Clinical grade Human Recombinant Interleukin-2: IL-2 Powder 22 MU VIAL (65483011607), storage in a -18 °C freezer where MU stands for medical units
Note: Alternatively, human recombinant IL-2 can be purchased from Life Technologies Corp, catalog number: PHC-0023. - Human Recombinant Interleukin-7: IL-7 Premium grade (Miltenyi Biotec, catalog number: 130-095-361), storage in a -18 °C freezer
- Efaviranz (EFV) (Cayman Chemical, catalog number: 14412), need to be re-suspended in IMDM media and diluted according to need of the experimental procedure
- Flow Monoclonal Antibodies
Note: You will use 5 μl of each antibody for staining. Use simple Flow staining protocol for staining the cells. This is just done to check for the purity of the resting T cells. Resting T cells are CD25-/CD69-/HLADR- cells.- Mouse Anti-human CD4-AF700 (BD Biosciences, catalog number: 557922)
- Mouse Anti-human CD3-V450 (BD Biosciences, catalog number: 560365)
- Mouse Anti-human CD25-APC (BD Biosciences, catalog number: 340939)
- Mouse Anti-human CD69-FITC (BD Biosciences, catalog number: 555530)
- Mouse Anti-human HLADR-PE (BD Biosciences, catalog number: 347367)
- Mouse Anti-human CD279-PeCy7 (BD Biosciences, catalog number: 561272)
Note: Isotype controls are used at the same concentration as the specific antibody. So, dilutions have to be determined according to the concentration of the matched antibody.- PE-CyTM 7 Mouse IgG1, κ Isotype Control (BD Biosciences, catalog number: 557646)
- FITC Mouse IgG1, κ Isotype Control (BD Biosciences, catalog number: 556649)
- V450 Mouse IgG1, κ Isotype Control (BD Biosciences, catalog number: 561504)
- Alexa Fluor® 700 Mouse IgG1, κ Isotype Control (BD Biosciences, catalog number: 557882)
- APC-H7 Mouse IgG1, κ Isotype Control (BD Biosciences, catalog number: 561427)
- PE Mouse IgG2a, κ Isotype Control (BD Biosciences, catalog number: 558595)
- Dye-eFluor 506 (Invitrogen, catalog number: 65-0866-14)
- DPBS (1x), Dulbecco’s Phosphate-Buffered Saline (Corning Cellgro, catalog number: 21-031-CV), store at 4 °C for use after opening bottle
- Dry Ice
- HBSS (1x), Hanks Balanced Salt Solution (Thermo Fischer Scientific, GibcoTM, catalog number: 141775-095), store at 4 °C for use after opening bottle
- IMDM (1x) (Thermo Fischer Scientific, GibcoTM, catalog number: 12440-053), store at 4 °C for use after opening bottle
- Lymphocyte separation medium (Corning, catalog number: 25-072-CV), store at 4 °C for use after opening bottle
- Penicillin/Streptomycin (Thermo Fischer Scientific, GibcoTM, catalog number: 15140-122), store at -18 °C freezer before opening bottle and then store at 4 °C for subsequent use
- RPMI 1640 (1x) with L-glutamine and 25 mM HEPES (Corning Cellgro, catalog number: 10-041-CV)
- 0.05% Trypsin EDTA (1x) (Thermo Fischer Scientific, GibcoTM, catalog number: 25300-120), store at -20 °C freezer before opening bottle and then store at 4 °C for subsequent use
- RoboSepTM Buffer (Stemcell Technologies, catalog number: 20104)
- Formalin
- 10% RPMI (see Recipes)
- 10% IMDM (see Recipes)
Note: This procedure can be done in a BSL2+ lab following safety procedures involving wearing a closed lab coat and double gloves. The blood and processing should all be done in a bio-safety hood with proper airflow.
Equipment
- Tissue culture flask vented caps, 70 ml (Falcon, catalog number: 353109)
- Tissue culture flask vented caps, 250 ml (Falcon, catalog number: 353110)
- Pipettes
- Rocker
- Incubator (Thermo Electron Corporation, NAPCO SERIES 8000 WJ, CO2 Incubator)
- Centrifuge: Sorvall Legend RT+ Centrifuge (Thermo Scientific)
- Freezer
- Luminometer (Promega GloMax Navigator, GM2000)
- LN2 freezer
- Magnets (Stem Cell Technologies, catalog number: 96290)
- Light Microscope
- 4 °C refrigerator
- Flow cytometer (BD, model: LSR II)
- Water Bath (Fisher Scientific, model: 2223)
- Haemocytometer 0.100 mm deep (Hausser Scientific)
Software
- FlowJo software (Tree Star)
- BD FACSDivaTM software (BD Biosciences)
- IUPMStats v1.0 Infection Frequency Calculator (Online) http://silicianolab.johnshopkins.edu/
- Microsoft Excel
Procedure
Notes:
- In order to get enough cells to keep the control for sets of experiment same, we leukophoresied control HIV-1 negative donors and also participants of the study, and froze those cells down at -140 °C.
- The TZA protocol may be adapted for fresh as well as frozen PBMCs.
- This procedure can be done in a BSL2+ lab following safety procedures involving wearing a closed lab coat and double gloves. The blood and processing should all be done in a bio-safety hood with proper airflow.
- Isolation of rCD4+ T cells from patients and controls
- Collect blood from participants in EDTA tubes. This is usually done in the hospital by nurses and then sent over for processing.
- Dilute the blood in DPBS (1:2). The total volume in a 40 ml tube should be 10 ml blood, 20 ml DPBS.
- Then use 10 ml of lymphocyte separation medium or ficoll to separate the layers of cells in the blood. This is done by putting the pipette with ficoll at the bottom of the 50 ml tubes and slowly pushing the liquid in and a layer of clear ficoll will be at the bottom of the tube. The total volume of liquid will not become 50 ml in the tube.
- Every reagent should be at room temperature before use.
- Spin this in a centrifuge at 2,000 rpm (448 x g) for 20 min to get distinct PBMC layer which will appear in between the ficoll layer and the DPBS and serum mixture. RBC will appear at the bottom of the tube.
- Collect the PBMCs layer and wash it again with DPBS spinning in a centrifuge at 1,000 rpm (112 x g) for 10 min.
- Discard the supernatant and collect the pellet (PBMCs). At this point you may either carry on with the rest of the procedure or freeze your cells in a freezing media (10% DMSO in FBS) and store them at -140 °C for long term storage.
- Count the cells recovered and then use resting CD4+ T cell Isolation kit to separate out the rCD4+ T cells from the PBMCs. It is always a good idea to not run less than 50 million cells at once since the yield in this kit is not high. It is a high purity but a low recovery kit. Also do not do two 5 min separation for less than 100 million cells, rather do one-time separation through the magnet. The details of the separation are listed in the protocol that is sent with the separation kit.
- Count the cells one more time before proceeding to the next step.
- You will have to take some cells out and stain for resting T cells. This is important since it helps you determine whether the cells selected are purely resting T cells.
- If we are working with rCD4+ T cells, you can expect about 5 to 15 million rCD4+T cells from thawing about 100 million PBMCs for patients and about 20-25 million rCD4+ T cells from 100 million normal control PBMCs. This will not change irrespective of fresh or frozen cells.
- For thawing frozen cells, put the frozen cells dropwise slowly while moving the tube in circular motion in a 15 ml tube containing 10 ml of 10% IMDM. Then spin the cells down at 1,000 rpm (112 x g) for 10 min to wash it off from the freezing media.
- Follow the same Steps A1-A9 for cells from negative control participants. For every experiment you will need to do a control PBMCs with which the cells will be compared.
- Assess the purity of resting T cells using Flow cytometry staining. (Figure 1)
- First, transfer cells (2 x 105 to 2.5 x 105) into a tube and centrifuge at 400 x g at 4 °C for 5 min and remove the supernatant.
- Perform the viability staining: resuspend the cell pellets with 250 μl of DPBS and 1 μl of Viability Dye-eFluor506. Incubate for 30 min at 4 °C.
- Wash the cells with DPBS and centrifuge the plate at 400 x g at 4 °C for 5 min and remove the supernatant.
- Resuspend the cell pellets first in 100 μl of DPBS and then put this in a mixture of the antibodies in 5 ml round-bottom polystyrene tubes with 5 μl of each antibody listed in Materials and Reagents and incubate for 30 min at 4 °C.
- Wash the cells at 400 x g at 4 °C for 10 min and remove the supernatant. Do this two times.
- Fix the cells in 1% Formalin to run your flow. You can run your flow in plates or tubes, but we prefer tubes.
- First gate on singlet population and them on lymphocytes. Follow this up by gating on the live population. In the live population of cells look specifically at the CD4+/CD3+ cells as these are the T cells.
- Then the rest of the gating is from the CD4+/CD3+ cells. Gate them for CD69, HLADR and CD25, which are all activation markers. PD1 is used for gating by us for determining cell death signal but it is not important to use if your follow up experiments do not require it.
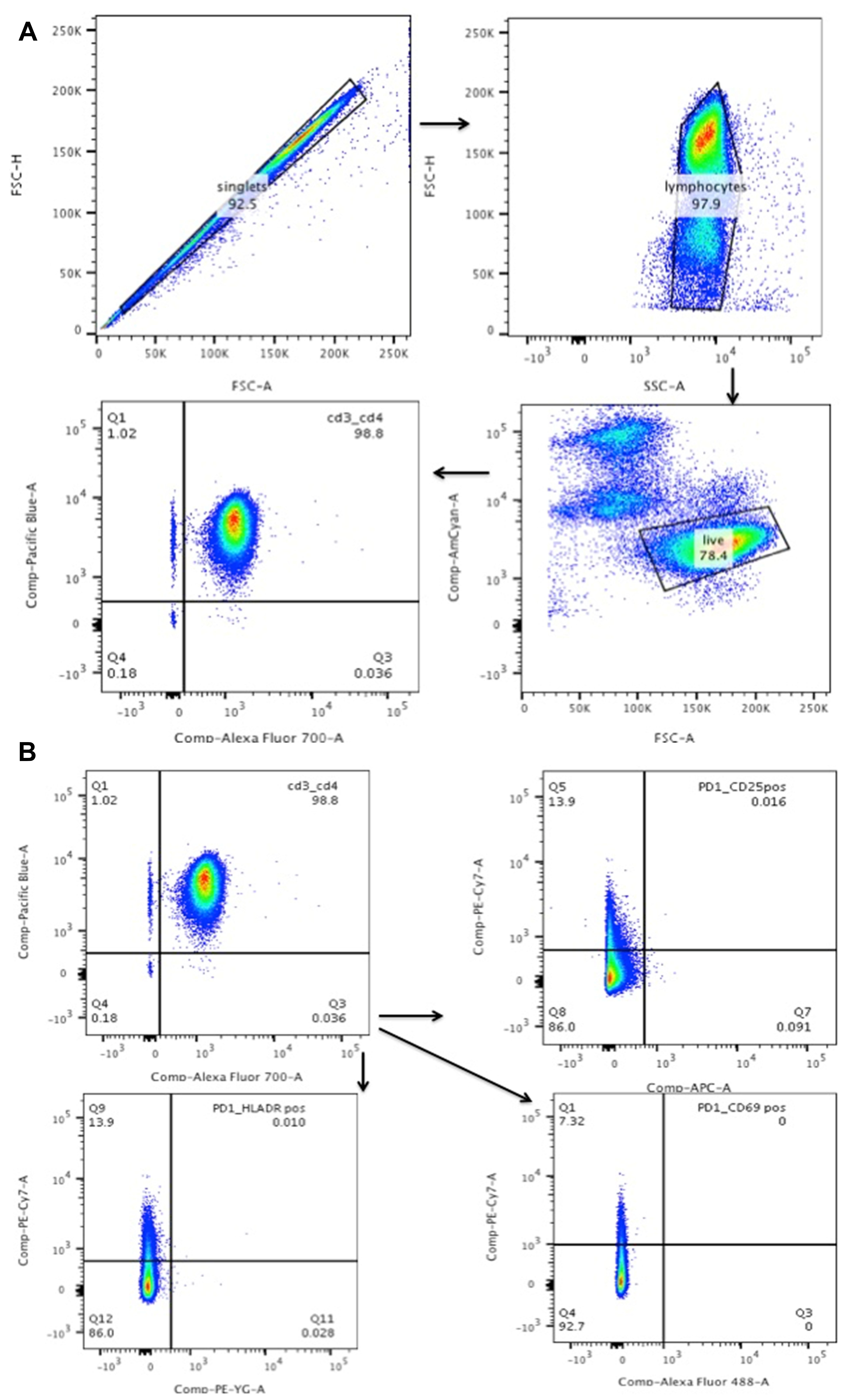
Figure 1. Purity of resting T cells: the resting T cells should be CD25/HLADR/CD69-CD3+/CD4+ cells. A. Gating on singlet population that is live and then to CD4+/CD3+ cells. B. Gating on CD3+/CD4+ cells to determine their activation status (CD25+/CD69+/HLADR+). - Activation of resting T cells
- For the next step you will need to first stimulate the cells for activation. Use Anti-CD3/CD28 Dynabeads in a concentration of 12.5 μl of the beads per million cells you put in the culture and they will stay in the culture together for 6 days till you do the next phase of the experiment.
- Wash the beads before using them for activation. Put beads in a 5 ml polystyrene tube, put RoboSepTM/EasySepTM buffer (1 ml) and put in magnet for 5 min. Then after decanting the liquid, resuspend the beads in 10% IMDM culture media exactly the amount you took out. For example, if you took out 100 μl of beads for 8 million cells, you wash the beads, then resuspend it back in 100 μl of 10% IMDM media and then out it in the final activation cocktail. (Figure 2).
- The culture for stimulation is usually placed in 1.0 million cells/ml of media. Each well in a 24-well plate will usually have 2 million resting T cells, 2 ml of 10% IMDM media and 25 μl of resuspended AntiCD3/CD28 Dynabead. After re-suspending the cells in media containing beads, put 300 nm of EFV per ml of the media to prevent cell-cell infection. Always prepare this fresh according to the amount of cell yield.
- For best stimulation, the cells can be concentrated in smaller wells (48-well plates with 1 million/ml cells in one well) and then transferred to a 24-well plate next day. Plate 2 million/2 ml of cells per well in a 24-well plate (Figure 2).
- After 24 h post activation, add 10 μl of 105 units/ml IL-2 and 10 μl of 105 units of IL-7per million cells as the secondary signaling cytokines to maintain the cells and help them proliferate. Do not remove the Anti-CD3/CD28 Dynabeads.
- The cultures need to be monitored every day and if there is overcrowding which is usually when the media becomes very yellow in color and the cell in each well is more than 4 million cells. Put more base media in the wells and split it into two wells in such cases.
- The base media during splitting should always contain IL-2, IL-7 (on Day 2) and EFV along with 10% IMDM. No beads are required during addition of media to the cells in culture (Figure 2).
- These cells stay in culture for a total of 6 days (Figure 2).
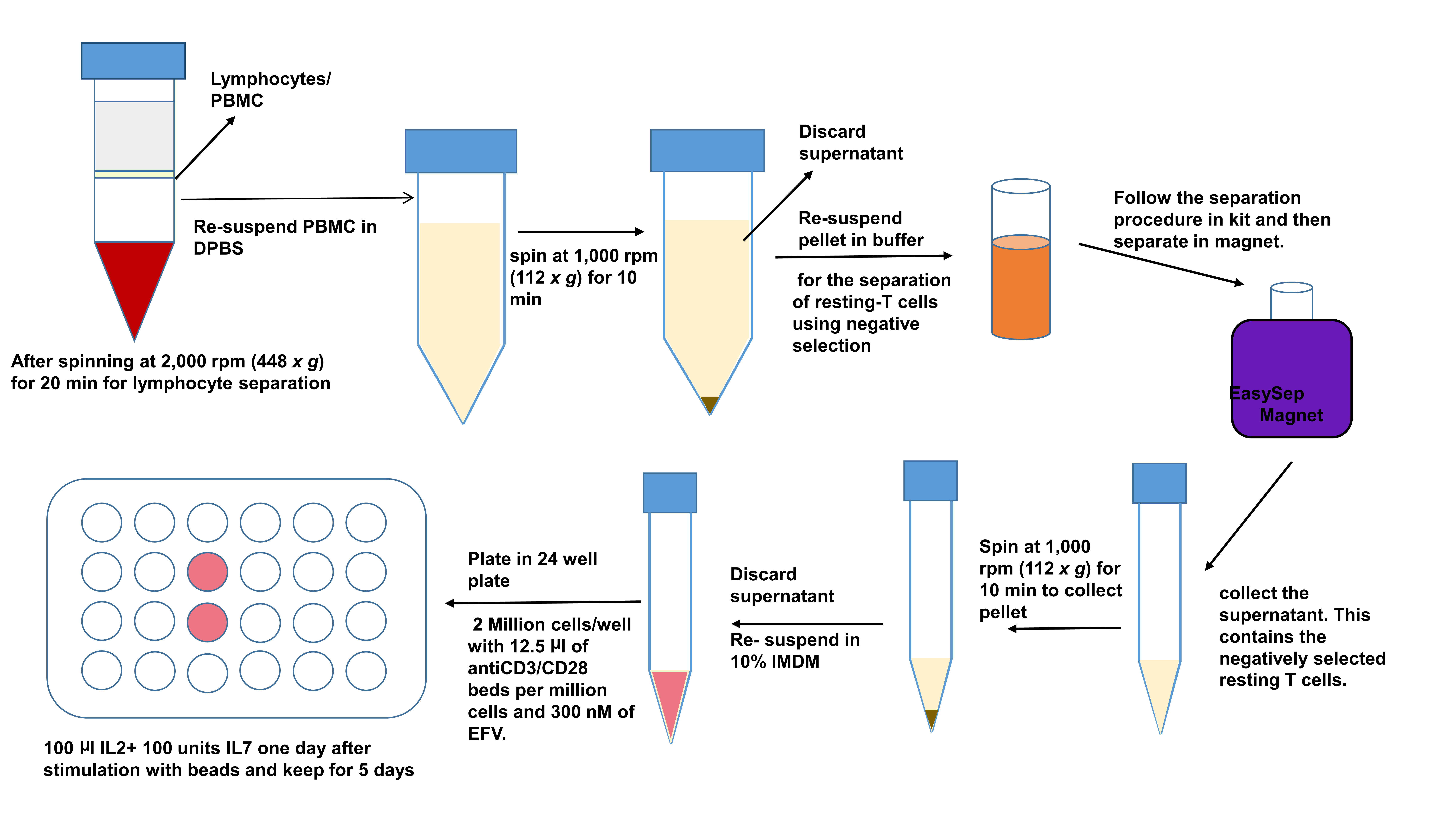
Figure 2. Processing latent cells for activation
- Harvesting, plating and reading Beta-gal activity of TZM-bl cells
- On Day 5 of the culture (one day before the T-cells are taken out of culture), seed 96-Well white PerkinElmer plates with 5 x 104 cells/well and rest of the plate with 3 x 104 cells/well of TZM-bl cells totaling 64-wells. Use 10% RPMI for media since TZM-bl cells grow in 10% RPMI. The total volume of media in each well should be 200 μl. Keep this overnight so that cells adhere to the bottom of wells by next day.
Note: Don’t maintain TZM-bl cells in culture for more than 3 weeks and split the cells every three days. When splitting, do not use more than 1 million/10 ml of 10% RPMI. TZM-bl cells grow in 10% RPMI in a 250 ml vented Falcon flask and need to be trypsinized when ready to split. When a frozen cell culture is started, make sure it is done in a 70 ml Falcon vented flask and then expanded over time. - On Day 6 from the start of culture, pull the activated T cells from the 24-well plates in 15 ml conical tubes and centrifuge at 1,000 rpm (112 x g) for 10 min to pellet the cells.
Note: You can save the supernatants for RNA if you want to compare the RNA yield of these activated latently infected cells. This can be frozen in -80 °C freezer. You will have to count the cells and wash them two more times with DPBS. You can also save 0.5 million cells for DNA quantification from each patient. For this, just count the cells, put them in Eppendorf tubes, spin them down at 1,000 rpm (112 x g) for 10 min and pellet the cells. Then directly just freeze these pellets in -80 °C freezer. This is used if you want to calculate the fraction of provirus induced for the particular participant. - With the rest of the activated T-cells, start to make a dilution series. Make a 4 fold dilution (1:3) starting from 1.25 x 105 cells/well in the highest dilution downward. (You can start as low as 6 x 104 cells/well on the top for this).
Note: If you are starting from 1.25 x 105 cells/well in the first row, each well in the top row (8 wells) should have 1.25 x 105 cells/well in 200 μl of 10% RPMI. The second row will have 1.25 x 105/4 cells in all eight wells and so on downwards for five more dilutions. This should give you a total of 6 dilutions (Figure 3). - After making the dilution series in 15 ml tubes, the media from the TZM-bl cell plates will have to be removed by aspiration before the patient cells are plated on it. Do not do it before you start to plate as it dries out the cells in the plate. The seventh row is usually not used and the eighth row is fresh media on the TZM-bl cells (Figure 3).
- All the dilutions will have to be in 10% RPMI.
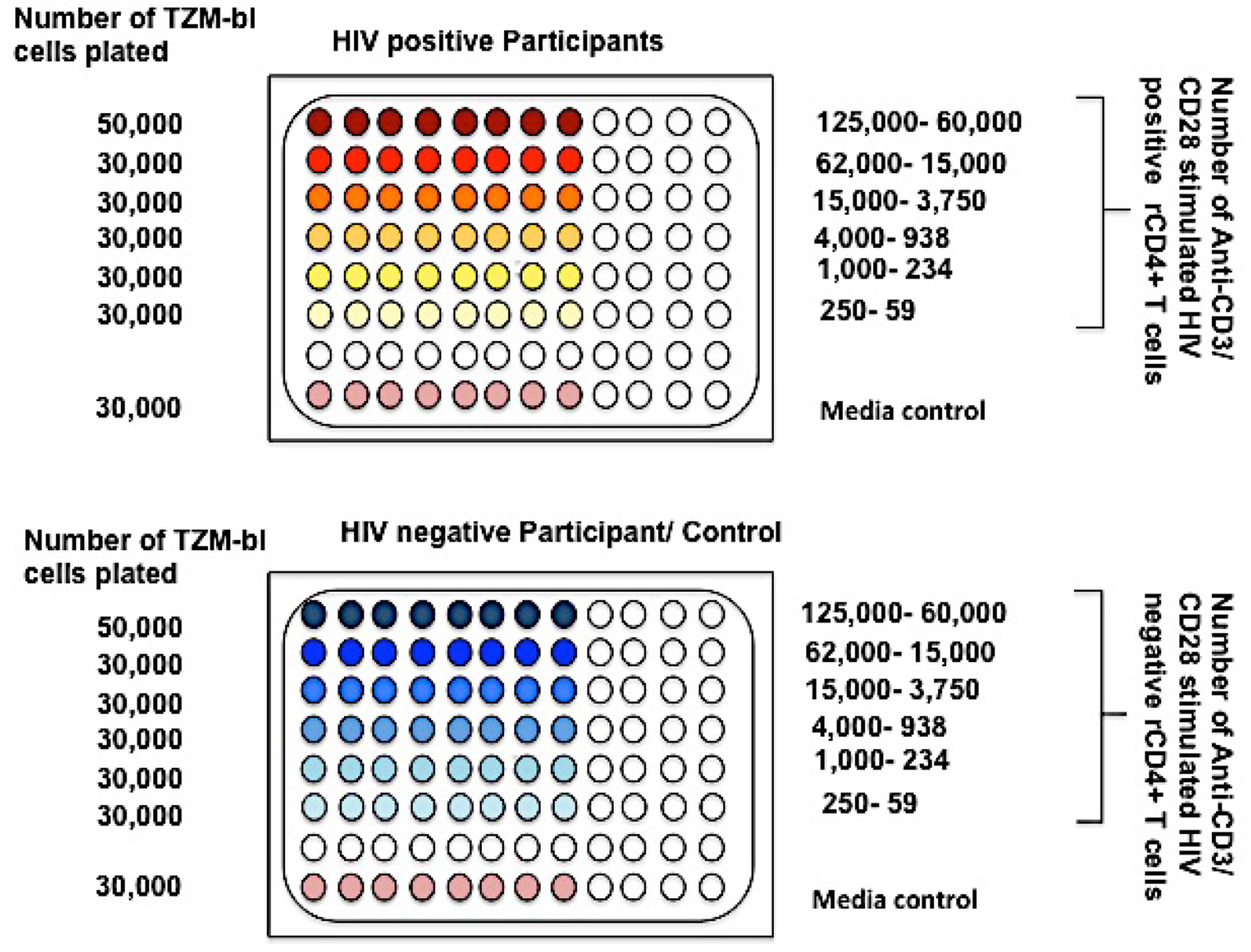
Figure 3. Plating cells for reading in 96 wells using TZM-bl cells - Keep this plate in a 37 °C incubator for 48 h after which it has to be read. This reading can be taken anywhere between 48 and 72 h.
- Take the plates out and aspirate the media out without disturbing the cells.
- Put back DPBS in the wells with multi-channel pipettes and lightly wash and take the washout and discard. Do this two times. Do not be very rigorous while washing as this could take the cells off from the plates.
- Put 100 μl of Promega Beta-Glo reagent on each well and incubate in the dark for 45 min-1 h. Try to do all these steps in very little light. This reagent is extremely sensitive to light.
- Then read in a Promega Glomax machine to get the light units following software prompt. This is an automated machine where you will have to select any of the cell-titer protocols, which will be able to read your plate. Then you can name and save the plate reads as excel files.
- On Day 5 of the culture (one day before the T-cells are taken out of culture), seed 96-Well white PerkinElmer plates with 5 x 104 cells/well and rest of the plate with 3 x 104 cells/well of TZM-bl cells totaling 64-wells. Use 10% RPMI for media since TZM-bl cells grow in 10% RPMI. The total volume of media in each well should be 200 μl. Keep this overnight so that cells adhere to the bottom of wells by next day.
- Analysis of readings and IUPM calculation
- After the reading is taken, transport the Excel file out to analyze these results. First, calculate a mean (average) of the bottom media control row and deduct this value from all the other individual readings in the plate. This will be your blank. This should be done for both the control HIV-1 negative plate as well as the HIV-1 positive patient plate. This will also assure that you ideally have similar conditions for both the test and control cells.
- In the control plate, which has the control HIV-1 negative T cells, calculate the average of each row with all the eight wells for each dilution. Do this for each dilution. Also, calculate the standard deviation for each dilution.
- Add the average + 2 times the standard deviation (avg. + 2 SD) for each dilutions. This will give you a value for each dilution in the control well based on which you will calculate the positive wells in the test plate.
- After you have the value of (avg. + 2 SD) for each dilution in the control plate, subtract it from each individual reading of wells for that particular dilution in the participant plate.
- This will give you some wells with positive and some with negative values. The positive values can vary from 0/8 to 8/8.
- Put these readings in the algorithm developed by Silicianos lab http://silicianolab.johnshopkins.edu/ (Rosenbloom et al., 2005) and calculate the Infectious unit per million cells or IUPM.
- For DNA copies/million cells, use a standard DNA q-PCR (Cillo et al., 2014) for integrated provirus in the cells and calculate the fraction of provirus that can be induced by the stimulation to form replication competent virus. This is calculated with the formula: fPVE = IUPM/DNA per million cells x 100.
Data analysis
- For the purity estimation of the resting T cells, samples are acquired on an LSR II flow cytometer using FACSDiVaTM software before putting the cells in culture for activation. Flow cytometry data are analyzed using FlowJo to gate, quantify, and analyze the resting T cell population.
- For the IUPM calculations, the maximum likelihood estimate was applied to determine the infectious unit per million (IUPM) cells for the TZA assay using online software, available at http://silicianolab.johnshopkins.edu/, developed by Rosenbloom et al. (2015).
Notes
During the entire protocol, you have to make sure that you are doing exactly the same things for another set of cells which are HIV-1 negative or control cells. If you do not have your controls, your experiments will not work. You will have to make sure even the control cells are stimulated the same way as the patient cells so that ideally you have similar conditions for both the test and control cells.
Recipes
- 10% RPMI
RPMI 1640
1x medium with 10% Animal Serum Complex and 1% Penicillin-streptomycin
Keep sterile and at room temperature during experiment and store at 4 °C - 10% IMDM
Mix 450 ml of IMDM w/L-glutamine with 50 ml FBS and 1% Penicillin-streptomycin (5 ml) from storage at -20 °C
This will make a final volume of 10% FBS in IMDM
This should always be stored at 4 °C for long term storage
Acknowledgments
This protocol was adapted from our publication (Sanyal et al., 2017). We thank the ex-members of the lab, Lori Caruso, Deena Ratner and Ming Ding (University of Pittsburgh, Pittsburgh) for their technical assistance; N. Sluis-Cremer for the DNA measurement and study design inputs; P. Tarwater (University of Texas, El Paso) and C. Shen (University of Pittsburgh, Pittsburgh) for statistical consultation and development of the assay statistics; W. Buchanan (University of Pittsburgh, Pittsburgh) for recruitment of the Multicenter AIDS Cohort Study participants for the study; and all the participants of the Pittsburgh portion of the MACS for donating blood for this study.
This work was supported by NIH grants R21AI138716 (Phalguni Gupta), U01-AI35041 (Charles R. Rinaldo.), R21-AI119117 (Nicolas P. Sluis-Cremer and NIH Fogarty training grant fellowship D43TW010039 (Phalguni Gupta).
Competing interests
The authors declare no conflict of interest.
Ethics
Blood was collected and handled according to protocols approved by the University of Pittsburgh institutional review board and was pulled from the patient in the hospital by nurses. The participants both HIV-1 negative and positive were recruited from the Multicenter AIDS Cohort Study.
Written consent was obtained from all the participants and the study was explained to them in simple terms before proceeding with it.
References
- Ananworanich, J. and Mellors, J. W. (2015). How much HIV is alive? The challenge of measuring replication competent HIV for HIV cure research. EBioMedicine 2(8): 788-789.
- Chun, T. W., Carruth, L., Finzi, D., Shen, X., DiGiuseppe, J. A., Taylor, H., Hermankova, M., Chadwick, K., Margolick, J., Quinn, T. C., Kuo, Y. H., Brookmeyer, R., Zeiger, M. A., Barditch-Crovo, P. and Siliciano, R. F. (1997). Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387(6629): 183-188.
- Cillo, A. R., Sobolewski, M. D., Bosch, R. J., Fyne, E., Piatak, M., Jr., Coffin, J. M. and Mellors, J. W. (2014). Quantification of HIV-1 latency reversal in resting CD4+ T cells from patients on suppressive antiretroviral therapy. Proc Natl Acad Sci U S A 111(19): 7078-7083.
- Finzi, D., Blankson, J., Siliciano, J. D., Margolick, J. B., Chadwick, K., Pierson, T., Smith, K., Lisziewicz, J., Lori, F., Flexner, C., Quinn, T. C., Chaisson, R. E., Rosenberg, E., Walker, B., Gange, S., Gallant, J. and Siliciano, R. F. (1999). Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 5(5): 512-517.
- Laird, G. M., Eisele, E. E., Rabi, S. A., Lai, J., Chioma, S., Blankson, J. N., Siliciano, J. D. and Siliciano, R. F. (2013). Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS Pathog 9(5): e1003398.
- Rosenbloom, D. I., Elliott, O., Hill, A. L., Henrich, T. J., Siliciano, J. M. and Siliciano, R. F. (2015). Designing and interpreting limiting dilution assays: General principles and applications to the latent reservoir for human immunodeficiency virus-1. Open Forum Infect Dis 2(4): ofv123.
- Sanyal, A., Mailliard, R. B., Rinaldo, C. R., Ratner, D., Ding, M., Chen, Y., Zerbato, J. M., Giacobbi, N. S., Venkatachari, N. J., Patterson, B. K., Chargin, A., Sluis-Cremer, N. and Gupta, P. (2017). Novel assay reveals a large, inducible, replication-competent HIV-1 reservoir in resting CD4+ T cells. Nat Med 23(7): 885-889.
- Siliciano, J. D., Kajdas, J., Finzi, D., Quinn, T. C., Chadwick, K., Margolick, J. B., Kovacs, C., Gange, S. J. and Siliciano, R. F. (2003). Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med 9(6): 727-728.
- Siliciano, J. D. and Siliciano, R. F. (2005). Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol Biol 304: 3-15.
- Siliciano, J. D. and Siliciano, R. F. (2004). A long-term latent reservoir for HIV-1: discovery and clinical implications. J Antimicrob Chemother 54(1): 6-9.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Sanyal, A., Rangachar, V. S. and Gupta, P. (2019). TZA, a Sensitive Reporter Cell-based Assay to Accurately and Rapidly Quantify Inducible, Replication-competent Latent HIV-1 from Resting CD4+ T Cells. Bio-protocol 9(10): e3232. DOI: 10.21769/BioProtoc.3232.
Category
Immunology > Immune cell isolation > Lymphocyte
Microbiology > Microbial cell biology > Cell-based analysis > Reporter assay
Cell Biology > Cell isolation and culture > Cell isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link