- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Northern Blot with IR Fluorescent Probes: Strategies for Probe Preparation
Published: Vol 9, Iss 8, Apr 20, 2019 DOI: 10.21769/BioProtoc.3219 Views: 11723
Reviewed by: Gal HaimovichPabitra Kumar SahooSwati MeghaAnca Savulescu

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Automated 384-well SYBR Green Expression Array for Optimization of Human Induced Pluripotent Stem Cell Differentiation
Max Y. Chen [...] Birgitt Schuele
Jun 5, 2023 2239 Views

Absolute Quantification of mRNA Isoforms in Adult Stem Cells Using Microfluidic Digital PCR
Shubhangi Das Barman [...] Antoine de Morree
Sep 5, 2023 1853 Views

Efficient circRNA Detection Using the Processive Reverse Transcriptase uMRT
Ruben Warkentin and Anna Marie Pyle
Oct 20, 2025 1392 Views
Abstract
Northern blot is a molecular biology technique that can detect, quantify, and determine the molecular weight of RNA. Recently, we published a protocol utilizing near-infrared (IR) fluorescent probes in Northern blot (irNorthern). Our method is as sensitive as other non-radioactive methods but is more straightforward and versatile. Additionally, we found that IR-labeled probes can be used to multiplex or detect different species of RNA at the same time. Here we describe three methods for generating an IR-labeled probe as well as how to perform irNorthern blot. In conclusion, our irNorthern protocol offers a convenient method for RNA detection.
Keywords: Northern blotBackground
Northern blot analysis is one of the foundational techniques available to biochemists and molecular biologists. While newer and more high-throughput techniques have been adopted, the Northern blot remains a powerful and adaptable method of analyzing RNA size and quantity simultaneously. In brief, DNA or RNA oligonucleotides labeled with radioactive phosphorus-32 (32P) are hybridized to a membrane crosslinked with RNA and subsequently detected using autoradiography (Alwine et al., 1977). Nonetheless, the use of 32P is subject to institutional regulation, and safety precautions must be observed during its use (Jones, 2005). Additionally, 32P probes have short lifespans increasing their cost for use. Alternative protocols using digoxigenin (DIG) conjugated to dNTP or NTP have been developed to replace 32P (Seibl et al., 1990). While successful, DIG probes require secondary detection by antibodies, more hands-on time and expensive reagents, deterring its wider adoption. Our recently published method, irNorthern, is straightforward and adaptable. It utilizes IR dyes which reduce auto-fluorescent background (Zarnegar et al., 2016; Miller et al., 2018). These IR dyes are conjugated with a dibenzocyclooctyne group (DBCO), which forms a covalent bond with the azide on the probes through copper-free click chemistry. To demonstrate its applications, we performed irNorthern to detect microRNA let-7a and small nuclear RNA U6 in total RNAs extracted from HCT116 wild-type and Drosha knockout cells (Kim et al., 2016). In addition, we provide details on how to generate three different azide-containing DNA or RNA probes for IR-dye labeling: a chemically synthesized DNA probe with an internal azide modification on dT, a DNA probe labeled with azide-dU by terminal transferase (TdT), and an RNA probe labeled with azide-U by T7 RNA polymerase.
Materials and Reagents
- Seal-Rite 1.5 ml microcentrifuge tube (USA Scientific, catalog number: 1615-5599)
- Hybridization tube (Fisher Scientific, catalog number: 13-247-300)
- Hybond N+ 30 cm x 3 M Roll (GE, catalog number: RPN303B)
- 50 ml centrifuge tubes (Genesee, catalog number: 21-106)
- 15 ml centrifuge tubes (Genesee, catalog number: 21-101)
- 1,250 µl pipet tips (USA Scientific, catalog number: 1112-1720)
- 0.1-10/20 µl pipet tips (USA Scientific, catalog number: 1110-3700)
- 200 µl pipet tips (USA Scientific, catalog number: 1110-1700)
- Whatman Grade 3 MM Chr blotting paper, sheet, 46 x 57 cm (GE, catalog number: 3030-917)
- 6-Well cell culture plates (Genesee Scientific, catalog number: 25-105)
- Paper towels (Highmark, catalog number 4497A1)
- Glass tray (Pyrex, catalog number: 1124847)
- Aluminum foil roll (Fisher, catalog number: 01-213-102)
- Sheet protectors (Office Depot, catalog number: 491694)
- HCT116 Parental cell line (Korean Collection for Type Cultures, catalog number: BP1230983)
- HCT116 Drosha KO #40 cell line (Korean Collection for Type Cultures, catalog number: BP1230984)
- let-7a irNorthern probe (5′-AACTATACAACCTACTACCTCA/iAzideN/A-3′)
Notes:- Probes are ordered from Integrated DNA Technology at 100 nmole scale, with one internal azide, and purified by HPLC.
- DNA probes are designed to be complementary to the target RNA. For miRNA targets, the probe is the full length (~22 nts) reverse complement of the mature miRNA. An internal azide is added close to the 3′ end with a terminal nucleotide as required for the synthesis.
- U6 irNorthern probe (5′-GCAGGGGCCATGCTAATCTTCTCTGTATCG/iAzideN/T-3′)
Note: Probe sequences for commonly used target RNAs can be found in the literature. When designing new DNA probes, we recommend use ~30 nt sequence targeting single-stranded regions of the RNA. - Terminal transferase (NEB, catalog number: M0315)
- 5-Azidomethyl-dUTP (Jena Biosciences, catalog number: CLK-084S)
- 5-Azido-C3-UTP (Jena Biosciences, catalog number: NU-157S)
- Agencourt AMPure XP beads (Beckman Coulter, catalog number: A63881)
- Isopropanol (J.T. Baker, catalog number: 908401)
- Ethanol (Decon Laboratories, catalog number: 2716)
- IRDye 680RD DBCO Infrared Dye, 0.5 mg (Li-Cor, catalog number: 929-50005)–light sensitive
- IRDye 800CW DBCO Infrared Dye, 5 mg (Li-Cor, catalog number: 929-55000)–light sensitive
- Urea (Sigma, catalog number: U5378)
- Acryl/Bis 19:1 (VWR, catalog number: 97064-990)
- Ammonium persulfate (APS) (AmericanBio, catalog number: AB00112)
- TEMED (N,N,N',N'-Tetramethylethylenediamine) (AmericanBio, catalog number: AB02020)
- Sodium chloride (AmericanBio, catalog number: AB01915)
- Sodium phosphate dibasic anhydrous (Fisher Scientific, catalog number: BP332-500)
- Potassium chloride (Sigma, catalog number: P9541)
- Sodium acetate (Sigma, catalog number: S7545)
- Trizma base (Sigma, catalog number: T1503)
- HEPES (Sigma, catalog number: H4034)
- Ethylenediaminetetraacetic acid (EDTA) (Sigma, catalog number: EDS)
- Sodium Dodecyl Sulfate (SDS) (AmericanBio, catalog number: AB01920)
- Formamide DI (AmericanBio, catalog number: AB00600-00500)
- Bromophenol blue (Sigma, catalog number: 114391)
- Xylene Cyanol FF (Sigma, catalog number: X4126)
- Sodium Citrate Dihydrate (Sigma, catalog number: W302600)
- Phenol/Chloroform/Isoamyl Alcohol (Fisher Scientific, catalog number: BP1752I-400)
- Chloroform (AmericanBio, catalog number: AB00350-01000)
- ExpressHyb hybridization solution (Clontech, catalog number: 636832)
- TRIzol (Ambion, catalog number: 15596018)
- SYBR Green I Nucleic Acid Gel Stain (Invitrogen, catalog number: S7563)
- A, C, U, G ribonucleotides (Thermo Fisher Scientific, catalog number: R0481)
- Spermidine trihydrochloride (Sigma, catalog number: 85578)
- RNase Inhibitor, Murine (NEB, catalog number: M0314L)
- Dithiothreitol (DTT) (AmericanBio, catalog number: AB00490-00025)
- Magnesium chloride hexahydrate (Sigma, catalog number: M2670)
- HYCLN McCoys 5A Media (Fisher Scientific, catalog number: SH30200)
- Fetal bovine serum (PAA Laboratories, catalog number: A15-201)
- Penicillin-Streptomycin (Thermo Fisher Scientific, catalog number: 15070063)
- RQ1 RNase-Free DNase (Promega, catalog number: M6101)
- 1x Phosphate Buffered Saline (PBS) (see Recipes)
- RNA Elution Buffer (see Recipes)
- 8 M urea 15% polyacrylamide gel (see Recipes)
- 8 M urea 6% polyacrylamide gel (see Recipes)
- 2x Formamide Loading Dye (see Recipes)
Equipment
- 16-Tube SureBeads magnetic rack (Bio-Rad, catalog number: 1614916)
- Vortex Genie 2 (Fisher Scientific, catalog number: 12-812)
- Micropipettes (Eppendorf, Research Plus Models)
- Roto-Bot rotating mixer (Benchmark Scientific, catalog number: R4045)
- Water-Jacketed CO2 incubator (Forma Scientific, catalog number: 3110)
- ThermoMixer C (Eppendorf, catalog number: 5382000015)
- NanoDrop OneC (Thermo Fisher Scientific, catalog number: ND-ONEC-W)
- Odyssey CLX Imaging System (Li-Cor)
- PowerPac HV High-Voltage power supply (Bio-Rad, catalog number: 1645056)
- PowerPac HC High-Current power supply (Bio-Rad, catalog number: 1645052)
- Owl HEP Series semidry electroblotting system (Thermo Fisher Scientific, catalog number: HEP-1)
- ImageQuant 300 imager (GE Healthcare, catalog number: 63-0056-52 )
- UV Transilluminator (Fotodyne, catalog number: 3-3000)
- UV Crosslinker, 254 nm (Spectroline, catalog number: XL1000)
- Innova 2000 platform shaker (New Brunswick, catalog number: M1190-0000)
- Hybridization oven (VWR, model number: 2720)
- Mini-PROTEAN Tetra vertical electrophoresis system (Bio-Rad, catalog number: 1658005)
Note: Although this system is designed for SDS-polyacrylamide gel electrophoresis (PAGE), we use it routinely to prepare and run Urea-PAGE for RNA separation. - Tabletop centrifuge with refrigeration (Eppendorf, catalog number: 5404000138)
- -20 °C freezer (GE, model number: FUM21DHRWW)
- -80 °C freezer (New Brunswick, model number: C760)
Software
- Image Studio (LiCor; free version available upon request at licor.com)
Procedure
- Preparation of IR-labeled probe
- Dissolve azide-containing DNA oligonucleotide with double distilled H2O (ddH2O). Final DNA concentration is 100 µM.
- Combine 2.5 nmole oligos with 50 nmole IRDye 680RD or 800CW DBCO in 1x phosphate buffered saline (PBS) in total volume of 50 µl.
Note: From this point on, the probe should be kept away from light. - Incubate reaction at room temperature for 6 h.
Note: Incubation was performed without agitation/shaking. We have also incubated overnight with no loss of yield. However, we have not determined the minimal incubation time for sufficient labeling. - Add 2 volumes of AMPure XP beads and 5.4 volumes of isopropanol; mix well by pipetting up and down 10 times.
- Incubate at room temperature for 7½ min. Remix, incubate for another 7½ min.
- Place beads on magnet stand. After beads settle, remove the supernatant.
- Wash twice with freshly prepared 85% ethanol.
- Elute in 100 µl of ddH2O; move the eluate to a new tube.
- Use NanoDrop to measure 1 µl and determine DNA concentration. Typical recovery efficiency is 70-90%.
- Alternative probe preparation I–prepare DNA probe using terminal transferase
- Prepare 30 µl terminal transferase (TdT) reaction in a 1.5 ml centrifuge tube with 1x Terminal Transferase Reaction Buffer, 0.25 mM CoCl2, 100 pmoles of DNA oligo (no internal azide), 300 pmoles of 5-Azidomethyl-dUTP, 20 U of TdT, and ddH2O.
Note: Reaction can be scaled up for preparing a large batch of probes. However, we recommend small scale TdT reaction with different probe:5-Azidomethyl-dUTP ratios to estimate labeling efficiency first. - Incubate reaction at 37 °C for 1 h in ThermoMixer C.
Note: Incubation was performed without agitation/shaking. - Follow Steps A4-A7 to purify the DNA using AMPure XP beads.
- Elute in 20 µl of ddH2O; move the eluate to a new tube.
- Measure DNA concentration by loading 1 µl on a NanoDrop.
- Label 19 µl of probe with 10 nmole IRDyes 680RD or 800CW DBCO in 1x PBS diluted in ddH2O.
Note: Make sure the molar ratio between IR dye and probe is greater than 10:1. - Incubate at room temperature for 6 h.
- Follow Steps A4-A7 to purify the probe using AMPure XP beads.
- Elute in 15 µl of ddH2O. Run entire IR-labeled oligo eluate on an 8 M urea 15% polyacrylamide gel and run 10 pmoles of unlabeled oligo.
Note: It is important to run unlabeled probe to estimate how many nucleotides have been added by TdT reaction. - Detect IR signal on Li-Cor Odyssey CLX Scanner (Figure 1A).
- Stain the gel with 1x SYBR Green I and visualize the signal in ImageQuant 300 Imager (Figure 1A).
- Match IR image and SYBR Green I staining image to determine the most prominently-labeled probes.
- Excise the gel slice containing the most prominently-labeled probes.
- Place gel pieces in a 1.5 ml centrifuge tube and elute overnight at room temperature in 600 µl 1:1 RNA Elution Buffer:Phenol/Chloroform/Isoamyl Alcohol with gentle rotation on a rotating mixer.
- Transfer the liquid to a new 1.5 ml tube.
- Vortex for 30 s and spin at 21,000 x g at 4 °C for 5 min.
- Transfer the aqueous phase to a new 1.5 ml centrifuge tube.
- Add 300 µl of chloroform, vortex for 30 s, and spin at 21,000 x g at 4 °C for 5 min.
- Transfer the aqueous phase to a new 1.5 ml centrifuge tube.
Note: We recommend adding 15 µg of glycogen to co-precipitate with the DNA and help visualize the pellet. - Add 1 ml cold 100% ethanol and incubate at -80 °C for 15 min.
- Spin at 21,000 x g for 30 min at 4 °C.
- Remove all the solution and air-dry the pellet for ~3 min or until dry.
- Resuspend RNA in 22 µl of ddH2O.
- Quantify 1 µl of RNA using NanoDrop.
- Prepare 30 µl terminal transferase (TdT) reaction in a 1.5 ml centrifuge tube with 1x Terminal Transferase Reaction Buffer, 0.25 mM CoCl2, 100 pmoles of DNA oligo (no internal azide), 300 pmoles of 5-Azidomethyl-dUTP, 20 U of TdT, and ddH2O.
- Alternative Probe Preparation II–generate RNA probes using T7 run-off Transcription
- Design and prepare PCR templates containing a T7 promoter to be used in a T7 run-off transcription reaction.
- Incubate overnight at 37 °C a 10 µl reaction containing 40 mM Tris-HCl pH 8.0, 25 mM NaCl, 2 mM Spermidine (HCl)3, 8 mM MgCl2, 1 mM ATP, 1 mM total UTP (3:1 ratio of UTP:5-azido-C3-UTP), 1 mM GTP, 1 mM CTP, 20 U Murine RNase Inhibitor, 10 mM DTT, and 4 U T7 RNA polymerase.
Note: Reaction can be scaled up for preparing a large batch of probes. However, we recommend several small scale T7 in vitro transcriptions with different UTP:5-azido-C3-UTP ratios to estimate transcription and subsequent IR dye labeling efficiency first. - Separate the in vitro transcribed RNAs using an 8 M urea 6% polyacrylamide gel and detect the RNA by UV shadowing, followed by excision of the gel slice containing the RNA.
- Place the gel slice in a 1.5 ml centrifuge tube and elute overnight at room temperature in 600 µl 1:1 RNA Elution Buffer:Phenol/Chloroform/Isoamyl Alcohol with gentle rotation on a rotating mixer.
- Follow Steps B15-B24 to extract and ethanol precipitate the RNA.
- Label RNA probe with IRDye 680RD or 800CW DBCO as described in Steps B6-B7.
- Purify by AMPure XP beads as described in Steps A4-A7.
- Elute in 15 µl of ddH2O. Run entire IR-labeled RNA eluate on an 8 M urea 6% polyacrylamide gel.
- Detect IR signal on Li-Cor Odyssey CLX Scanner (Figure 1B).
- Stain the gel with 1x SYBR Green I and visualize the signal with ImageQuant 300 Imager (Figure 1B).
- Excise the labeled probes.
- Place gel pieces in a 1.5 ml centrifuge tube and elute overnight at room temperature in 600 µl 1:1 RNA Elution Buffer:Phenol/Chloroform/Isoamyl Alcohol with gentle rotation on rotating mixer.
- Follow Steps B15-B24 to extract and ethanol precipitate the RNA probe.
- TRIzol extraction of RNA
- Culture HCT116 and HCT116 Drosha knockout cells in a 6-well plate in McCoy’s 5A Media with 10% fetal bovine serum and 1% penicillin and streptomycin.
- When confluent, remove media and rinse cells with 1x PBS. Collect cells in a 1.5 ml microcentrifuge tubes and pellet at 1,000 x g for 3 min at 4 °C.
- Remove bulk of the supernatant, briefly resuspend cells in the residual (~20-30 µl) 1x PBS.
Note: If TRIzol is added to a dry cell pellet, the cells will not lyse completely. - Add 1 ml of TRIzol to each cell pellet collected from one well and vortex for 30 s.
- Add 200 µl of chloroform, vortex for 30 s and spin at 12,000 x g for 15 min at 4 °C.
- Transfer the aqueous phase (about 550 µl) to a new 1.5 ml microcentrifuge tube.
- Precipitate the RNA by adding equal volume (550 µl) of isopropanol and incubate at room temperature for 10 min.
- Spin at 21,000 x g for 20 min at 4 °C, a white pellet should form at the bottom of the tube.
- Remove the supernatant and wash the pellet with 1 ml of cold 75% ethanol.
- Spin at 21,000 x g for 5 min at 4 °C.
- Remove supernatant, air-dry pellet for ~3 min, and resuspend RNA in 20 µl of ddH2O.
- Mix 1 µl of RQ1 DNase with 5 µl 10x DNase RQ1 buffer and 24 µl ddH2O and add mix to RNA.
- Incubate reaction at 37 °C for 30 min.
- Bring volume to 300 µl with ddH2O and add 300 µl of Phenol/Chloroform/Isoamyl Alcohol.
- Follow Steps B16-B24 to extract and ethanol precipitate the total RNA. Glycogen could be omitted.
- Urea-PAGE, transfer, and Northern blot
- Mix 15-20 µg of total RNA sample with equal volume of 2x Formamide Loading Dye, heat samples at 95 °C for 3 min, immediately place on ice, and separate RNA on an 8 M urea 15% polyacrylamide gel.
- Transfer the gel at 300 mAmps for 1 h to Hybond N+ membrane using Owl HEP Series semidry electroblotting system.
Note: Alternatively, RNAs separated by Urea-PAGE could be transferred using wet-transfer system (RNA: a laboratory manual, Chapter 3, Protocol 11). If RNAs were separated on an agarose gel, traditional capillary transfer method should be used (RNA: a laboratory manual, Chapter 3, Protocol 9). - Briefly dry membrane on a paper towel and crosslink membrane twice using 254 nm UV crosslinker at 120 mJ/cm2.
- In the hybridization oven, pre-hybridize the membrane in a hybridization tube with 10 ml ExpressHyb hybridization solution for 30 min at 30 °C.
- Remove the pre-hybridization solution and hybridize overnight at 30 °C with 10 pmoles IR probe diluted in 10 ml ExpressHyb hybridization solution.
Notes:- If different probes have been labeled with different IRDyes, they can be combined together to detect different RNAs simultaneously.
- Hybridization temperature should be adjusted for different probes, ~20 °C below calculated Tm. We recommend a hybridization temperature of 30 °C for miRNAs. When using RNA probes, we typically perform hybridization at 65 °C overnight, with no loss of signal due to temperature.
- Collect diluted probe in a 15 ml centrifuge tube and store at -20 °C.
Note: Probes can be reused several times without significant loss of signal. - Place membrane in a clean glass tray and add 2x SSC (Saline-sodium citrate)-0.1% SDS Wash Buffer to wash the membrane.
Note: Submerge the membrane under ~1 cm solution. - Shake at 110 rpm for 10 min at room temperature.
Note: Washing temperature can be adjusted for different probes, depending on the Tm between the probe and the target. - Perform second wash with 1x SSC-0.1% SDS Wash Buffer for 10 min at room temperature with shaking at 110 rpm.
Note: If higher washing stringency is desired, continue with washing buffer containing lower SSC concentration and higher SDS concentration. - Briefly tap dry the membrane with paper towels.
- Image on Li-Cor Odyssey CLX Scanner to detect emission at 700 nm and/or 800 nm (Figure 1C).
- To strip the probes off the membrane; place membrane in a glass tray, pour microwave boiled 0.1x SSC-1% SDS-40 nM Tris-HCl pH 8.0, shake at 110 rpm for 10 min at room temperature.
- Repeat strip procedure (Step E12), dry membrane with paper towels, and store in sheet protector.
Note: Membrane can be stored for years and re-probed later.
Data analysis
Data is analyzed using the Image Studio software. Users can adjust brightness and contrast without changing the original image. The software also allows for the quantitation of signal, although we did not do it in this protocol. For users who want to multiplex, the software simultaneously scans at two separate wavelengths and overlays the images, generating two independent but perfectly superimposed images.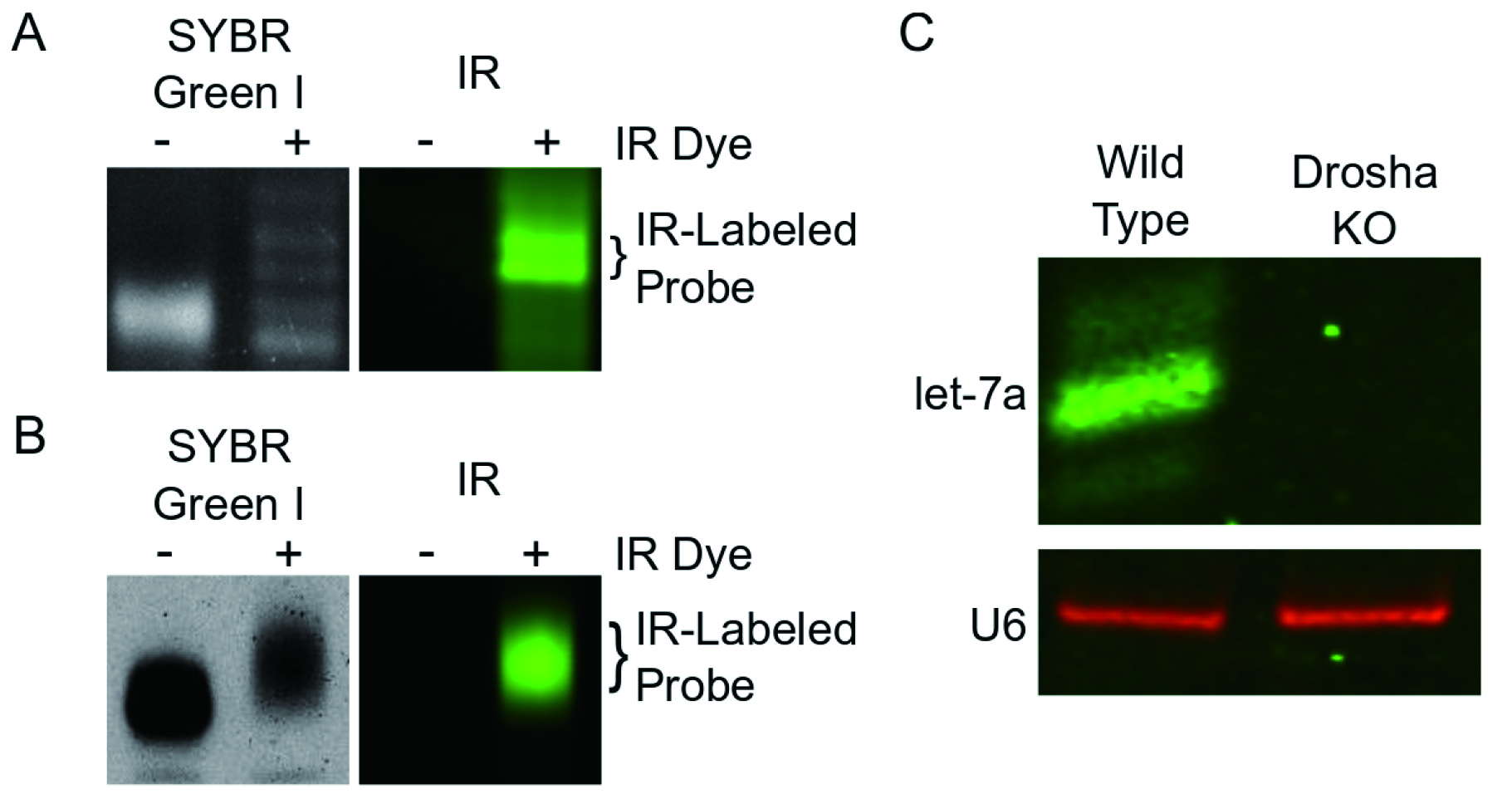
Figure 1. Alternative generation of IR probes and detection of let-7a and U6 using IR probes. A. DNA IR-probes generated from terminal transferase reactions and separated on an 8 M urea 15% polyacrylamide gel. The gel was scanned on a Li-Cor Odyssey CLX Scanner and then stained with SYBR Green I before being visualized using a UV transilluminator. The bands containing the desired IR-labeled probe are indicated by brackets. B. RNA IR-probe generated by T7 in vitro transcription and separated on an 8 M urea 6% polyacrylamide gel. The gel was scanned on the Li-Cor Odyssey CLX Scanner, stained with SYBR Green I, and then visualized on a UV transilluminator. The IR-labeled RNA was excised. C. irNorthern simultaneously detects miRNA let-7a and small nuclear RNA U6. Total RNA from HCT116 wild type and Drosha Knockout (KO) cells were extracted using TRIzol, separated on an 8 M urea 15% polyacrylamide gel, and transferred to a membrane. irNorthern probes for let-7a and U6 were hybridized overnight and the membrane was visualized using the Li-Cor Odyssey CLX Scanner. Canonical miRNAs (i.e., let-7a) require Drosha for processing and are not expressed in Drosha KO cells (Kim et al., 2016). U6 is utilized as a loading control. Images in Figures 1A and 1B were originally published in RNA under Miller et al., 2018 and can be found at https://rnajournal.cshlp.org/content/24/12/1871.
Notes
- When working with IR dyes, it is important to keep stocks away from light using aluminum foil wrapped tubes or amber tubes. Additionally, we have found that IRDye 800CW-labeled probes are more sensitive than 680RD-labeled probes. Therefore, we suggest for multiplexing, using the 800CW-labeled probes for less abundant targets.
- RNA probes generated in Procedure C cannot be completely stripped from the membrane after hybridization to the target RNAs.
Recipes
- 1x Phosphate Buffered Saline (PBS) (pH 7.4)
137 mM NaCl
10 mM Na2HPO4
2.7 mM KCl - RNA Elution Buffer
300 mM NaOAc
25 mM Tris-HCl, pH 8.0 - 8 M urea 15% polyacrylamide gel
5 g Urea
2 ml 5x TBE (Tris-Borate-EDTA)
625 µl ddH2O
*Microwave until the solution is warm (~5-8 s)
3.75 ml 40% Acrylamide
*Rotate until urea completely dissolved
52 µl 10% APS
10 µl TEMED - 8 M urea 6% polyacrylamide gel
5 g Urea
2 ml 5x TBE
2.875 ml ddH2O
*Microwave until the solution is warm (~5-8 s)
1.5 ml 40% Acrylamide
*Rotate until urea completely dissolved
52 µl 10% APS
10 µl TEMED - 2x Formamide loading Dye
80% Formamide
0.1% Bromophenol Blue
0.1% Xylene Cyanol
20 mM EDTA
20 mM Tris-HCl, pH 8.0
Acknowledgments
The Xie Lab is supported by the National Institute of Health (R00-CA190886 and R35-GM128753 to M.X.), University of Florida Research Opportunity Seed Fund (to M.X.), and Thomas H. Maren Foundation (Junior Investigator Fund F013372 to M.X.).
Competing interests
The authors declare no conflict of interest or competing interest.
References
- Alwine, J. C., Kemp, D. J. and Stark, G. R. (1977). Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc Natl Acad Sci U S A 74(12): 5350-5354.
- Jones, C. G. (2005). A review of the history of U.S. radiation protection regulations, recommendations, and standards. Health Phys 88(2): 105-124.
- Kim, Y. K., Kim, B. and Kim, V. N. (2016). Re-evaluation of the roles of DROSHA, Exportin 5, and DICER in microRNA biogenesis. Proc Natl Acad Sci U S A 113(13): E1881-1889.
- Miller, B. R., Wei, T., Fields, C. J., Sheng, P. and Xie, M. (2018). Near-infrared fluorescent Northern blot. RNA 24(12): 1871-1877.
- Rio, D.C., Hannon, G.J., Ares Jr., M., and Nilsen, T.W. (2011). RNA: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press. ISBN: 978-0-879698-91-1.
- Seibl, R., Holtke, H. J., Ruger, R., Meindl, A., Zachau, H. G., Rasshofer, R., Roggendorf, M., Wolf, H., Arnold, N., Wienberg, J. and et al. (1990). Non-radioactive labeling and detection of nucleic acids. III. Applications of the digoxigenin system. Biol Chem Hoppe Seyler 371(10): 939-951.
- Zarnegar, B. J., Flynn, R. A., Shen, Y., Do, B. T., Chang, H. Y. and Khavari, P. A. (2016). irCLIP platform for efficient characterization of protein-RNA interactions. Nat Methods 13(6): 489-492.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Fields, C., Sheng, P., Miller, B., Wei, T. and Xie, M. (2019). Northern Blot with IR Fluorescent Probes: Strategies for Probe Preparation. Bio-protocol 9(8): e3219. DOI: 10.21769/BioProtoc.3219.
Category
Molecular Biology > RNA > RNA detection > Northern Blot
Biochemistry > RNA > miRNA tagging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











