- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantification of Hepatitis B Virus Covalently Closed Circular DNA in Infected Cell Culture Models by Quantitative PCR
Published: Vol 9, Iss 7, Apr 5, 2019 DOI: 10.21769/BioProtoc.3202 Views: 8283
Reviewed by: David PaulGundeep KaurAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Triplet-primed PCR and Melting Curve Analysis for Rapid Molecular Screening of Spinocerebellar Ataxia Types 1, 2, and 3
Mulias Lian [...] Samuel S. Chong
Jun 20, 2023 1248 Views

Development of a Rapid Epstein–Barr Virus Detection System Based on Recombinase Polymerase Amplification and a Lateral Flow Assay
Yidan Sun [...] Chao Shen
Dec 5, 2024 1619 Views

Rapid Plasmid-Free Generation of Recombinant Positive-Strand RNA Viruses That Use IRES-Mediated Translation Using an Expansion of the Circular Polymerase Extension Reaction (CPER)
Hirotaka Yamamoto [...] Takausuke Fukuhara
Apr 20, 2025 2071 Views
Abstract
Persistence of the human hepatitis B virus (HBV) requires the maintenance of covalently closed circular (ccc)DNA, the episomal genome reservoir in nuclei of infected hepatocytes. cccDNA elimination is a major aim in future curative therapies currently under development. In cell culture based in vitro studies, both hybridization- and amplification-based assays are currently used for cccDNA quantification. Southern blot, the current gold standard, is time-consuming and not practical for a large number of samples. PCR-based methods show limited specificity when excessive HBV replicative intermediates are present. We have recently developed a real-time quantitative PCR protocol, in which total cellular DNA plus all forms of viral DNA are extracted by silica column. Subsequent incubation with T5 exonuclease efficiently removes cellular DNA and all non-cccDNA forms of viral DNA while cccDNA remains intact and can reliably be quantified by PCR. This method has been used for measuring kinetics of cccDNA accumulation in several in vitro infection models and the effect of antivirals on cccDNA. It allowed detection of cccDNA in non-human cells (primary macaque and swine hepatocytes, etc.) reconstituted with the HBV receptor, human sodium taurocholate cotransporting polypeptide (NTCP). Here we present a detailed protocol of this method, including a work flowchart, schematic diagram and illustrations on how to calculate “cccDNA copies per (infected) cell”.
Keywords: Hepatitis B VirusBackground
The Hepatitis B virus (HBV), a DNA virus belonging to the family Hepadnaviridae, is a human pathogen persisting in approximately 240 million people globally. HBV infection leads to higher risks of liver cirrhosis and hepatocellular carcinoma (Liang et al., 2015). At present, chronic HBV infection is not curable as current treatments do not eradicate the replicative reservoir, covalently closed circular (ccc)DNA (Levrero et al., 2016). In in vitro infected hepatocytes, cccDNA is formed by cellular repair of relaxed circular duplex (rc)DNA, the genomic form in virions (Guo et al., 2007; Long et al., 2017; Schreiner and Nassal, 2017). cccDNA serves as the viral template for pregenomic RNA which becomes encapsidated into nucleocapsids and is reverse transcribed to generate rcDNA and double-stranded linear (dsl)DNA by the viral polymerase (Bartenschlager and Schaller, 1992; Tu et al., 2017). Of note, albeit the reconstitution of hepatoma cells with the receptor human sodium taurocholate cotransporting polypeptide (NTCP), high excess of inoculated virions with high multiple genome equivalents (mges) (mges > 100) is required to achieve moderate infection rates (e.g., > 20% in HepG2NTCP cells) (Qu et al., 2018). This raises the problem that cccDNA-containing samples taken early after inoculation with HBV contain large amounts of rcDNA from input virions.
Therefore, adequate methodologies for absolute or relative quantification of cccDNA are required. They are divided into two categories: (I) hybridization after separation by gel electrophoresis (Southern blot) and (II) PCR amplification (Li et al., 2017). Southern blot is still a gold standard but not very sensitive, and demands multiple experimental processes and a high-copy load of cccDNA (> 2 x 106 copies using 32P-radioactive isotope/digoxigenin/biotin probe; > 1 x 104 copies using branched DNA technique) (Yu et al., 2015). Therefore, hybridization-based methods are complicated, time-consuming and not practical for a large number of samples (e.g., > 20) to be analyzed in parallel. PCR methods include real-time quantitative (q)PCR, nested qPCR (Xu et al., 2011), digital-droplet PCR (Mu et al., 2015), and rolling circle amplification (Margeridon et al., 2008). Real-time qPCR is the fastest and the most robust method for almost all laboratories. However, unlike Southern blot which gel electrophoretically separates cccDNA, PCR methods are not strictly specific, especially when rcDNA and other HBV replicative intermediates are present in excess, such as in in vitro infection (e.g., MGE > 300), even when cccDNA-specific primer pairs are used (Nassal, 2015; Qu et al., 2018).
To solve this problem, we have developed a qPCR assay using validated cccDNA selective primer pairs and a digestion step by T5 exonuclease, which removes cellular DNA and all HBV intermediates via its exonuclease activity targeting free ends of rcDNA and dslDNA but leaves cccDNA intact (Qu et al., 2018). This assay allowed fast and specific quantification of cccDNA within one working day as shown in Figure 1 (2 h of total DNA extraction; 1.5 h of T5 exonuclease reaction and 2 h of qPCR), accurate calculation of “cccDNA copies/(infected) cell” (Figure 2) and drug efficacy testing on cccDNA levels (Figure 3). The method also provided quantitative judgment on whether cccDNA is formed in new cell models and identified low amount of cccDNA in in vitro infection of non-primate hepatocytes (Lempp et al., 2016 and 2017). This protocol is adapted from Qu et al. (2018) and herein more detailed information on this qPCR quantification after T5 exonuclease digestion is included, and different primer pairs are compared to address the applicability of HBV genotypes. Taken together, this protocol will facilitate studies on cccDNA and help clinicians, technicians and graduate students to analyze cccDNA in samples derived from in vitro infection.
Materials and Reagents
- Pipette tips (Neptune, 1,000 μl, 200 μl, 20 μl, 10 μl, DNase-/RNase-free & Biozym, premium tips 1000 μl, 200 μl, 10 μl)
- 1.5 ml microcentrifuge tube (Sarstedt AG & Co.KG, SafeSeal tube, catalog number: 72.706)
- 0.2 ml microcentrifuge tube (Greiner Bio-one, Sapphire PCR tube, catalog number: 683271)
- Hard-Shell PCR plates 96-well, thin-wall (Bio-rad, catalog number: HSP9601)
- Primary human hepatocytes (PHH) (obtained from Hannover medical school or prepared in University Hospital Heidelberg) (Possible commercial vendors of PHH are Lonza, Biopredic, BioIVT, ThermoFisher, however susceptibility to HBV may vary), HepaRGNTCP and HepG2NTCP cell lines (Qu et al., 2018)
- Plasmid pSHH2.1 (Cattaneo et al., 1983), available upon request from the corresponding author
- Primers:
Primer p1040: 5’-GTGGTTATCCTGCGTTGAT-3’
Primer p1996: 5’-GAGCTGAGGCGGTATCT-3’
p1578: 5’-CCGTGTGCACTTCGCTTCA-3’
p1867: 5’-GCACAGCTTGGAGGCTTGA-3’
p1583: 5’-TGCACTTCGCTTCACCT-3’
p2301: 5’-AGGGGCATTTGGTGGTC-3’ - Probe
Probe p1085: 5’-FAM-AGTTGGCGAGAAAGTGAAAGCCTGC-TAMRA-3’ - Trypsin [0.25% Trypsin-EDTA (1x)] (Invitrogen, Gibco, catalog number: 25200-056), 4 °C
- NucleoSpin Tissue kit (Macherey-Nagel, catalog number: 740952.250)
- Buffer T1 (Macherey-Nagel, catalog number: 740940.25)
- Proteinase K (Macherey-Nagel, catalog number: 740506), -20 °C
- Buffer B3 (Macherey-Nagel, catalog number: 740920)
- Ethanol absolute (VWR chemicals, catalog number: 20821.330)
- Buffer BW (Macherey-Nagel, catalog number: 740922)
- Buffer B5 Concentrate (Macherey-Nagel, catalog number: 740921)
- T5 exonuclease (New England Biolabs, catalog number: M0363), -20 °C
- NEBuffer 4 (New England Biolabs, catalog number: B7004), -20 °C in aliquots
- PerfeCTa qPCR Toughmix (Quanta Biosciences, catalog number: 95112-012), -20 °C in aliquots
- SYBR Green Supermix (Bio-Rad, catalog number: 172-5121), -20 °C in aliquots
- Myrcludex B (Bachem), available upon request from the corresponding author, -80 °C in aliquots
- Recombinant human Interferon-α-2a (PeproTech, catalog number: 300-02AA), -80 °C in aliquots
- Human Interferon-α-2a (PBL Assay Science, catalog number: 11100-1), -80 °C in aliquots
- Nuclease-free water (B. Braun Melsungen, Aqua ad iniectabilia Braun)
- NaCl (Carl Roth GmbH, catalog number: 9265.2)
- KCl (Sigma-Aldrich, catalog number: 31248)
- Na2HPO4·2H2O (Sigma-Aldrich, catalog number: 04272)
- KH2PO4 (Sigma-Aldrich, catalog number: P9791)
- Tris-base (Carl Roth GmbH, catalog number: 4855.2)
- Dimethyl sulfoxide (Sigma-Aldrich, catalog number: 1.02950)
- Fetal bovine serum gold (PAA Laboratories GmbH, catalog number: A15-151), -20 °C
- Penicillin/Streptomycin (Thermo Fisher Scientific, catalog number: 15140-122), 4 °C
- L-glutamine (Thermo Fisher Scientific, catalog number: 25030-024), 4 °C
- MEM non-essential amino acids (Thermo Fisher Scientific, catalog number: 11140-035), 4 °C
- Recombinant insulin (Sigma-Aldrich, catalog number: 91077C-1G), -20 °C in aliquots
- Hydrocortisone 21-hemisuccinate sodium salt (Sigma-Aldrich, catalog number: H4881), -20 °C in aliquots
- Culture medium (see Recipes)
- 10x PBS buffer (see Recipes)
- 5 mM Tris-HCl (see Recipes)
Equipment
- Pipettes (Gilson P1000, P200, P20; Eppendorf P2.5)
- Cell counter (Bio-Rad, TC20TM Automated Cell Counter, catalog number: 1450102)
- Thermomixer (Eppendorf, ThermoMixer C, catalog number: 5382000015)
- Microcentrifuge (Thermo Fisher Scientific, HeraeusTM PicoTM 21 Centrifuge, catalog number: 75002553)
- Thermocycler (Labrepco, Biometra T3000 Thermocycler 48, catalog number: 050-723)
- qPCR thermocycler (Bio-Rad, C1000 TouchTM Thermal cycler with 96-well Fast Reaction Module, catalog number: 1851196)
Software
- CFX96 Real-time System
(Bio-Rad, http://www.bio-rad.com/en-us/product/cfx96-touch-real-time-pcr-detection-system) - CFX ManagerTM Software
(Bio-Rad, http://www.bio-rad.com/en-us/sku/1845000-cfx-manager-software?ID=1845000)
Procedure

Figure 1. Working flowchart of this protocol. Susceptible cells (PHH, HepaRGNTCP, HepG2NTCP) infected with high mges (> 300) of HBV are lysed. A. Lysates are incubated at 70 °C in the presence of proteinase K and later loaded on silica column and eluted. B. T5 exonuclease (5 units/1 h) removes host genomic DNA (black) and HBV replicative intermediates (rcDNA, dslDNA, etc.) (green) and preserves cccDNA (red) intact. C. cccDNA is amplified by p1040/p1996 and detected by probe p1085.
- HBV DNA extraction from in vitro infected hepatocytes (Figure 1A)
- For infection performed in a 24-well plate (culture area: 2 cm2), trypsinize and resuspend infected PHH, differentiated HepaRGNTCP or HepG2NTCP cells (infection procedure see Ni and Urban, 2017) in 1 ml of culture medium. Take 10 μl of the resuspension for cell count by a cell counter (see Equipment). Calculate total cell numbers.
Notes:- At confluency, cells in one well of a 24-well plate approximately correspond to 180,000 PHH, 200,000 HepaRGNTCP and 500,000 HepG2NTCP cells.
- When smaller or larger plates are used, volumes of the respective buffers given in the protocol should be adjusted according to the respective culture area (cm2) of a well.
- Spin down the cells (900 x g, 5 min) at room temperature. Wash cells with 1 ml of PBS twice. Spin down the cells and carefully aspirate PBS (Optional: freeze cell pellet at -20 °C for long-term storage). For DNA extraction, manufacturer’s manual of the NucleoSpin Tissue kit (740952.250) is adapted with a minor modification regarding the incubation time in Step A2 (1 h instead of 10~15 min) and the elution volume in Step A6 (50 μl instead of 100 μl). Add 200 μl of lysis buffer T1, pipet up and down and incubate at room temperature for 10 min. Add 25 μl of proteinase K and 200 μl of lysis buffer B3, vortex and incubate the lysate at 70 °C for 1 h with shaking (500 rpm).
Note: After proteinase K digestion, it is not necessary to re-centrifuge the lysates. Immediately perform the next step. - Add 210 μl of ethanol (96%-100%) to the sample and vortex vigorously for a few seconds. Remove buffer from the lid by a short centrifugation and apply the samples to the column. Centrifuge (11,000 x g, 1 min) at room temperature. Discard flow-through.
- Add 500 μl of wash buffer BW. Centrifuge (11,000 x g, 1 min) at room temperature. Discard flow-through.
- Add 600 μl of wash buffer B5 (pre-add indicated volume of ethanol to the Buffer B5 Concentrate) to the column. Centrifuge (11,000 x g, 1 min) at room temperature. Discard flow-through. Repeat this step once.
- Remove residual ethanol from the column (13,000 x g, 1 min). Place the column into a 1.5 ml microcentrifuge tube. Add 50 μl of pre-warmed (70 °C) elution buffer. Incubate at room temperature for 1 min. Centrifuge (11,000 x g, 1 min). Freeze eluted DNA samples at -20 °C until use or analyze them immediately.
Notes:- Elution buffers are nuclease-free water or 5 mM Tris-HCl (pH = 8.0). Do not use buffers with EDTA that may inhibit enzymatic activity in the next step.
- DNA concentration in eluates is 200-1,000 ng/μl.
- Preserve at least 10 μl of the sample without T5 exonuclease digestion for quantification of β-globin (internal standard) for normalization use.
- For infection performed in a 24-well plate (culture area: 2 cm2), trypsinize and resuspend infected PHH, differentiated HepaRGNTCP or HepG2NTCP cells (infection procedure see Ni and Urban, 2017) in 1 ml of culture medium. Take 10 μl of the resuspension for cell count by a cell counter (see Equipment). Calculate total cell numbers.
- T5 exonuclease hydrolysis of total DNA elutes (Figure 1B)
- Assemble the reaction components in a 0.2 ml microcentrifuge tube following the table as shown:
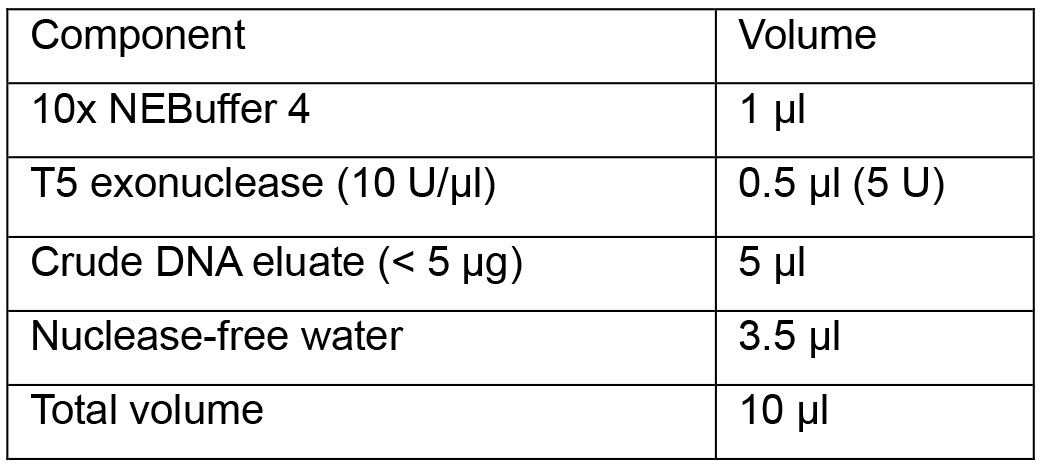
Note: Do not exceed unit and incubation time of T5 exonuclease since overdigestion leads to partial loss of cccDNA.
- Incubate the reaction at 37 °C for 1 h and inactivate the enzyme at 70 °C for 20 min in a thermocycler with heat-lid supply.
- Proceed quantification by real-time qPCR or freeze the products at -20 °C until use.
- Assemble the reaction components in a 0.2 ml microcentrifuge tube following the table as shown:
- Quantification of cccDNA relative to human β-globin (single-copy gene) by real-time qPCR (Figure 1C)
- Quantification of cccDNA
- Dilute primers and probe to 10 μM:

FAM: 6-carboxyfluorescein; TAMRA: 6-carboxytetramethylrhodamine - Prepare the following qPCR reactions in a 96-well Hard-Shell PCR plate:
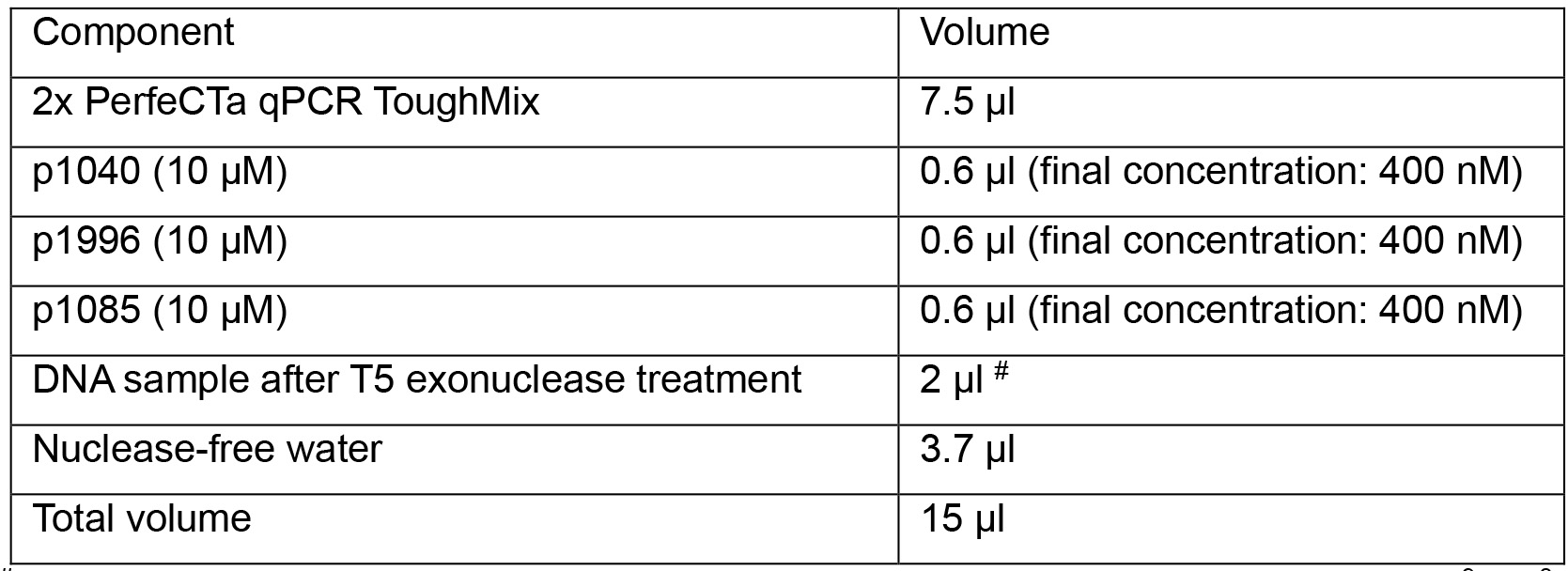
#for absolute quantification, load 2 µl of each 1:10 serial-diluted plasmid (pSHH2.1: 109, 108, 107, 106, 105, 104, 103, 102 copies/μl) as templates on the same plate. - Run qPCR program below:
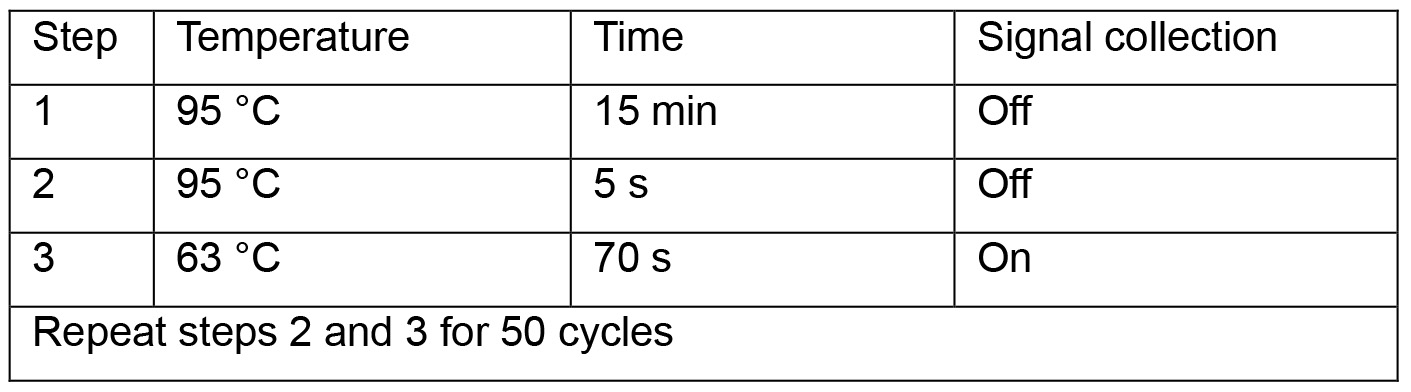
- Read absolute cccDNA copy numbers in the CFX ManagerTM Software. Calculate cccDNA copies per well.
- Dilute primers and probe to 10 μM:
- Quantification of human β-globin prior to T5 exonuclease digestion
- Dilute primers at 10 μM:
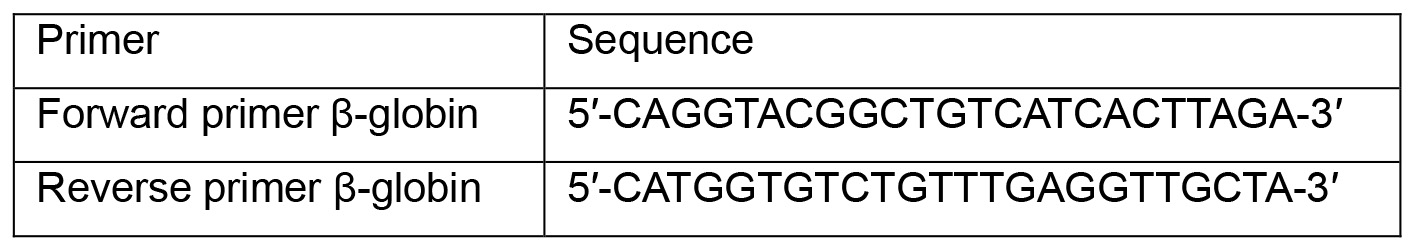
- Prepare qPCR reactions in a 96-well plate:
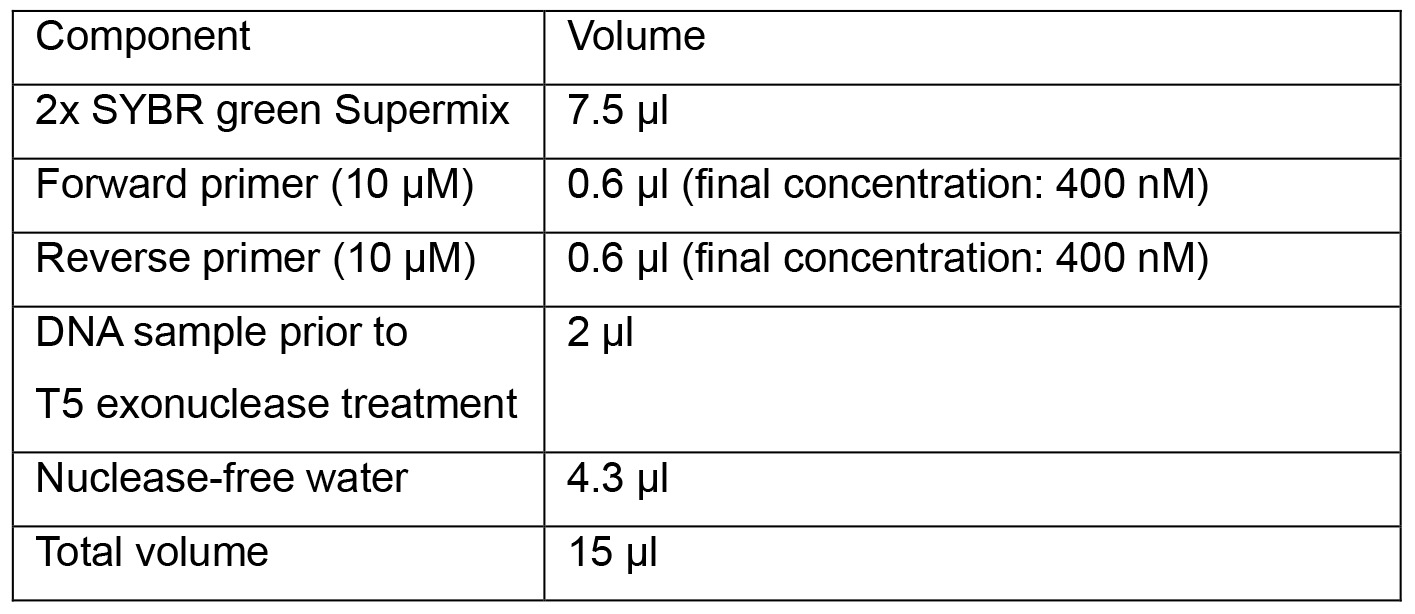
- Start qPCR program below:
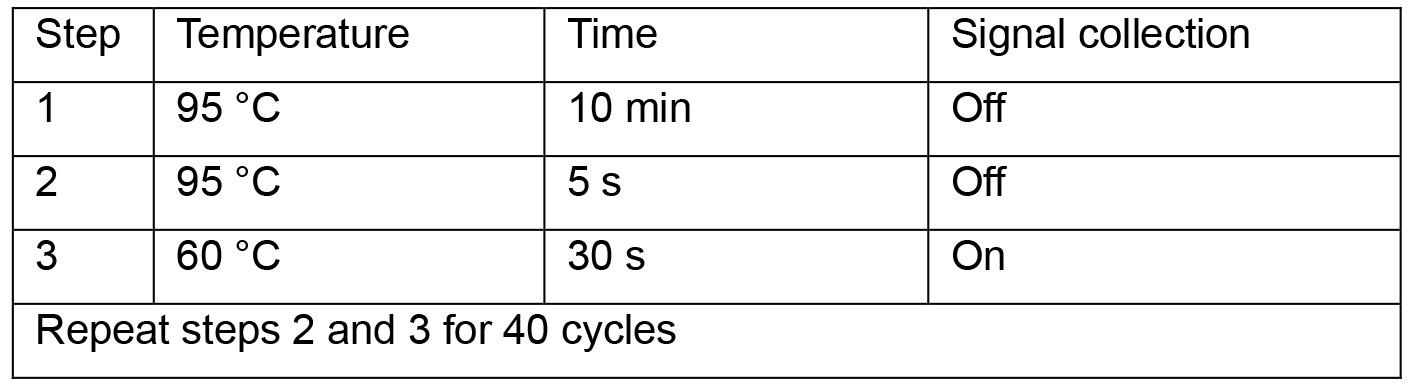
- Read levels of human β-globin. Normalize cccDNA copies per well using values of human β-globin. Calculate cccDNA copies per cell (copies per well divided by cell numbers per well). For instance, if cell numbers in a well of 24-well plate are 4.2 x 105 (as determined in Procedure A1) and cccDNA copies per well are 8.52 x 105 (as determined in Procedure C), cccDNA copies per cell are 2.03.
- Dilute primers at 10 μM:
- Quantification of cccDNA
Data analysis
cccDNA copy number per cell or infected cell is calculated according to Figure 2. The cccDNA level in in vitro infected cells can be determined similarly as two examples shown in Figure 3 and Qu et al., 2018. Note that to discriminate signals from the inoculum, we highly recommend using an entry inhibitor (e.g., Myrcludex B) as a control. Routinely Myrcludex B control gives a value below 0.05 copy per infected cell.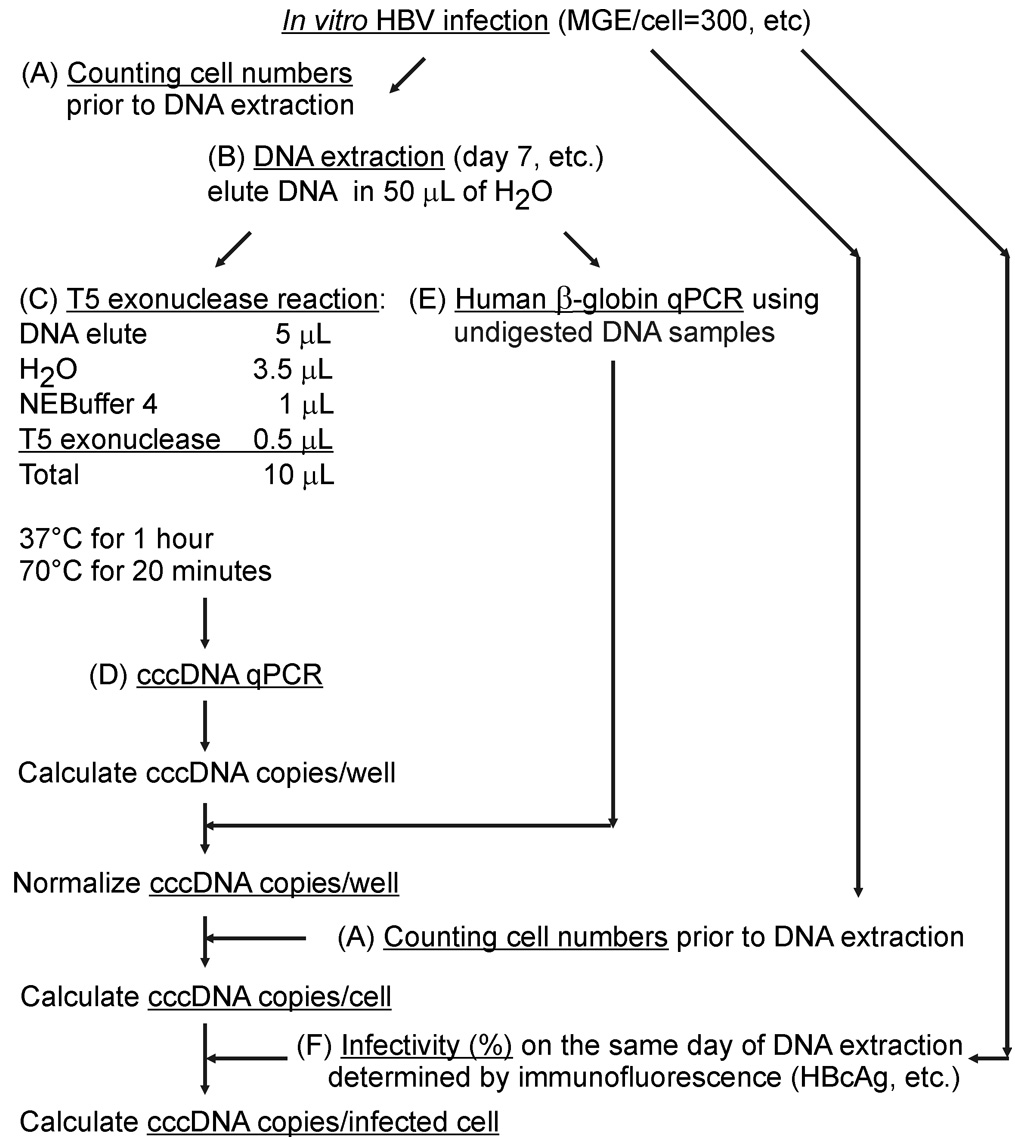
Figure 2. Schematic diagram of the calculation of absolute cccDNA copy numbers. A. On the day of experiment, cell number in the well is determined. B. Total DNA samples are extracted as suggested in Procedure. C. Here shows a proper T5 exonuclease digestion as shown in Procedure. D. Absolute cccDNA copies and E level of human β-globin are quantified, respectively as shown in Procedure. “cccDNA copies/cell” is “cccDNA copies/well” divided by “cell numbers”. Optional: F. If calculation of copies per infected cell is required, additional wells have to be arranged in parallel on the same plate during the infection. On the day of DNA extraction, cells in the wells are fixed and subjected to an immunofluorescence assay (HBcAg visualization using DAKO B0586 antibody, etc.) to determine infectivity (%: number of HBcAg positive cells divided by number of total cells) (Qu et al., 2018). “cccDNA copies/infected cell” is the “cccDNA copies/cell” divided by “infectivity (%)”.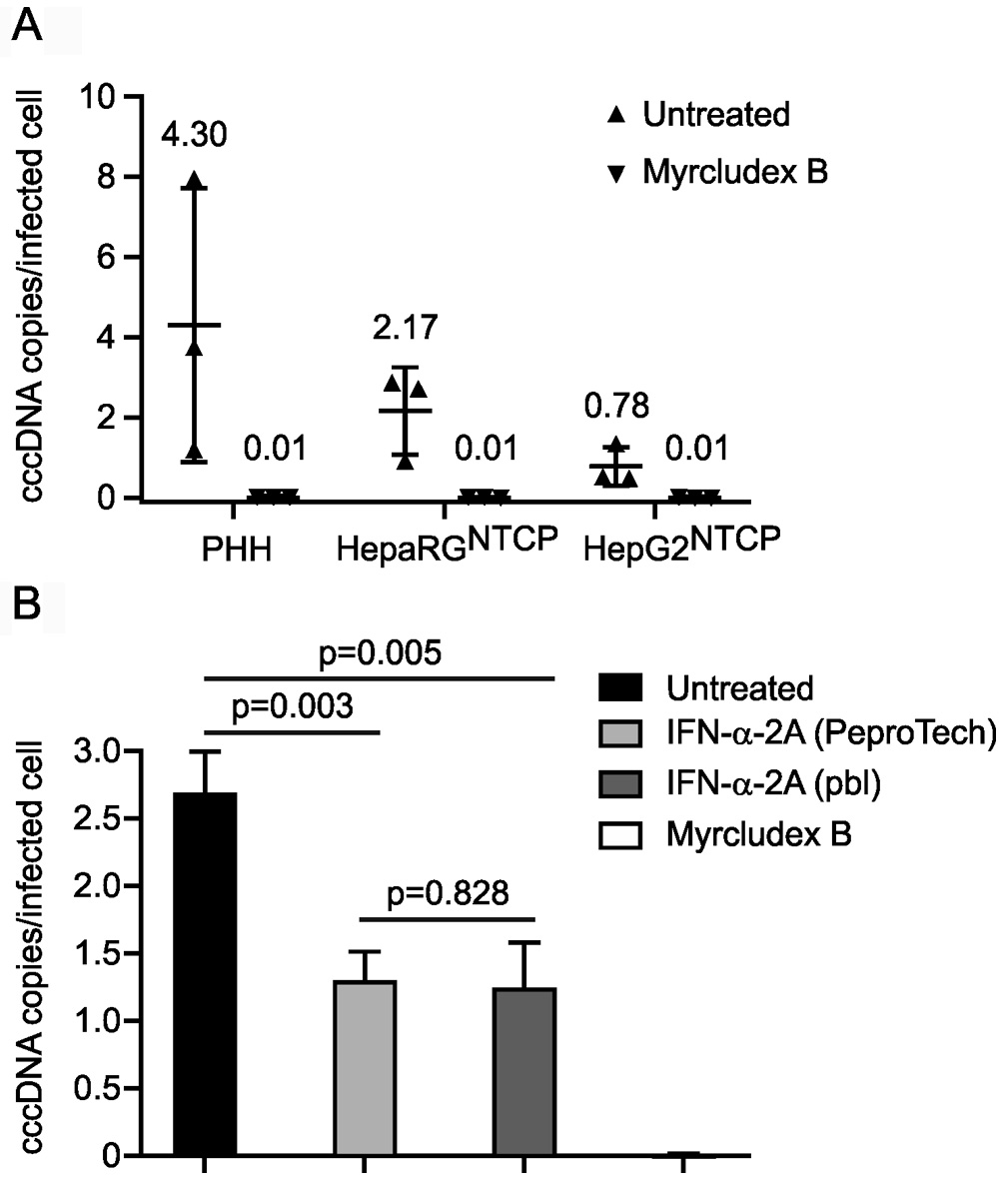
Figure 3. cccDNA levels in three in vitro. models and upon IFN-α-2a and Myrcludex B treatment. A. PHH, differentiated HepaRGNTCP and HepG2NTCP cells were infected with HBV at mges/cell of 500. Myrcludex B (1 μM), an entry inhibitor blocking cccDNA formation, was co-administered with HBV inoculum during infection. On Day 7 post infection, cccDNA copies per infected cell were determined. Data shown in triangles were collected from three independent experiments. B. HepG2NTCP cells were infected with HBV at mges/cell of 500 with mock treatment (untreated), co-treated with Myrcludex B (1 μM) during infection, or co- and post-treated with IFN-α-2A at 100 ng/ml purchased from PeproTech and PBL Assay Science. On Day 7 post infection, cccDNA copies per infected cell were analyzed. Statistics: P < 0.01 untreated versus IFN-treated; P = 0.828 PeproTech versus PBL.
Notes
The primer pair (pp1040-1996) is specific for the formed cccDNA in genotype D. Genotype D is the HBV genome in HepAD38 cell line. Users should consider this issue if they try patient-derived serum to perform in vitro infection using this protocol since HBV in patients can be any genotype and pp1040-1996 does not bind the formed cccDNA of other genotypes.
Genbank accession numbers of other genotypes: A (HE974370.1); B (AB540582.1); C (AB540584.1); E (HE974384.1); F (HE974369.1); G (AP007264.1); H (AB846650.1). pp1578-1867 and pp1583-2301 (Qu et al., 2018) allowing quantification of eight HBV genotypes (A-H) are suggested below. (Table 1)
Table 1. List of primer mismatches to HBV genotypes. The numbers of base-pair mismatch of each forward and reverse primers to eight HBV genotypes are summarized below. Green squares: perfect binding; yellow squares: one base-pair mismatch still allowing template binding; red squares: not binding.
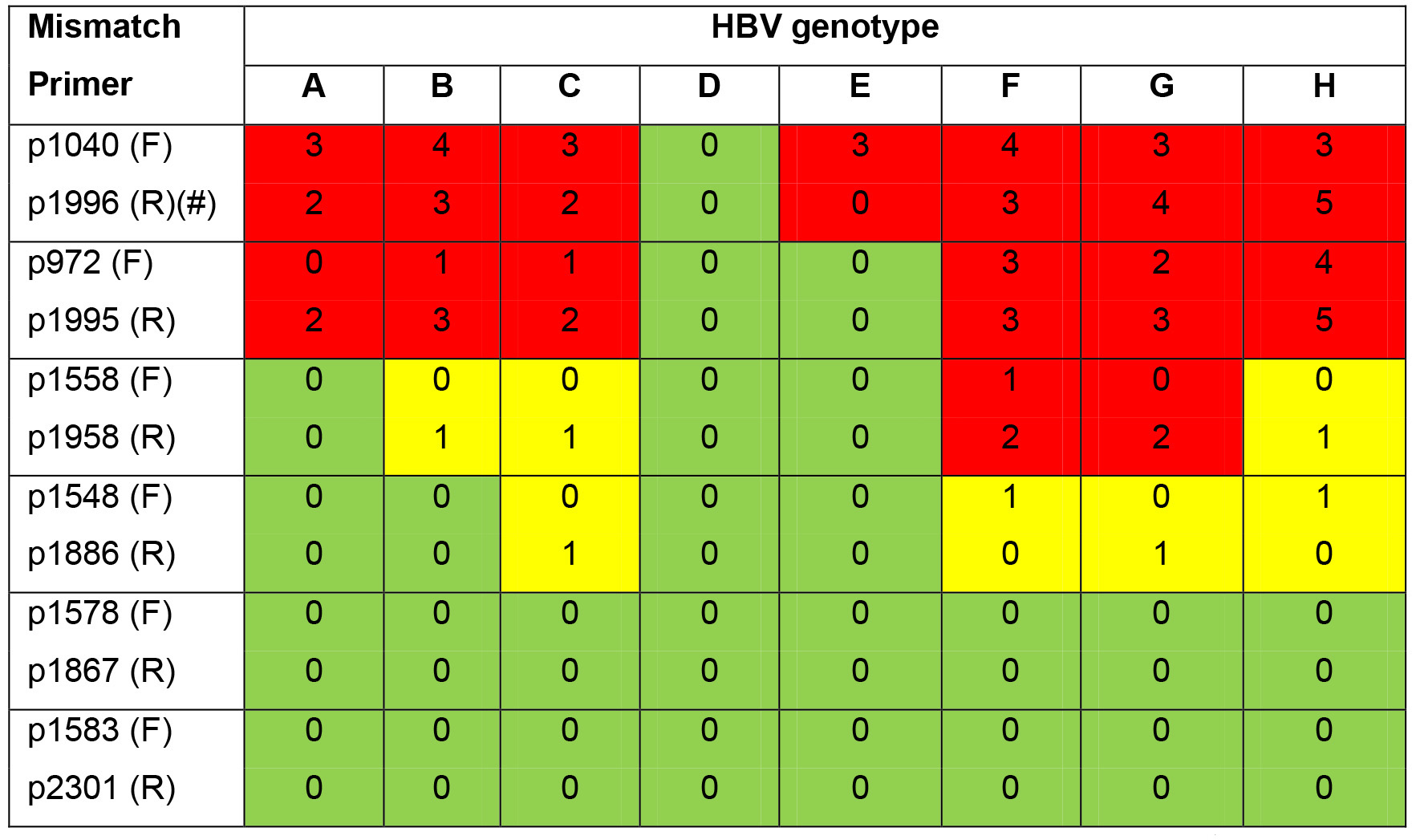
F: forward primer; R: reverse primer. #: The probe of this protocol is not pan-genotypic and has mismatch number as shown: A(1), B(2), C(0), D(0), E(0), F(6), G(1), H(4).
Recipes
- Culture medium
For HepG2NTCP cells: DMEM supplemented with 10% fetal calf serum (heat-inactivated), 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine, 1% MEM non-essential amino acids. For infection, add 2% dimethyl sulfoxide
For PHH and HepaRGNTCP cells: William’s medium E supplemented with 10% fetal calf serum (heat-inactivated), 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine, 1% MEM non-essential amino acids, 5 μg/ml recombinant insulin, 50 μM hydrocortisone. For differentiation and infection, add 1.5% dimethyl sulfoxide - 10x PBS
Dissolve 80 g of NaCl, 2 g of KCl, 11.5 g of Na2HPO4·2H2O and 2 g of KH2PO4 in 800 ml of H2O, adjust pH to 7.4, refill H2O to 1 L - Tris-HCl (1 M)
Dissolve 121.14 g of Tris-base in 800 ml of H2O, adjust pH to 8.0 and refill H2O to 1 L
Acknowledgments
This protocol was adapted from Qu et al., 2018. We herein acknowledge financial supports by Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)–Project number 272983813–SFB/TRR179 (B.Q. and S.U.) and German Center for Infection Research (DZIF) TTU Hepatitis projects 05.704 (S.U.). We thank Pascal Mutz (DKFZ, Germany) and Florian WR Vondran (MHH, Germany) for providing PHH. We are grateful to Xue Li for critical reading of this manuscript.
Competing interests
Prof. Dr. Stephan Urban, the corresponding author, holds patents and intellectual property on Myrcludex B.
Ethics
Following written informed consent of the patients, PHH were isolated from liver specimens obtained after partial hepatectomy.
References
- Bartenschlager, R. and Schaller, H. (1992). Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. Embo j 11(9): 3413-3420.
- Cattaneo, R., Will, H., Darai, G., Pfaff, E. and Schaller, H. (1983). Detection of an element of the SV40 late promoter in vectors used for expression studies in COS cells. Embo j 2(4): 511-514.
- Guo, H., Jiang, D., Zhou, T., Cuconati, A., Block, T. M. and Guo, J. T. (2007). Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis B virus: an intermediate of covalently closed circular DNA formation. J Virol 81(22): 12472-12484.
- Lempp, F. A., Qu, B., Wang, Y. X. and Urban, S. (2016). Hepatitis B virus infection of a mouse hepatic cell line reconstituted with human sodium taurocholate cotransporting polypeptide. J Virol 90(9): 4827-4831.
- Lempp, F. A., Wiedtke, E., Qu, B., Roques, P., Chemin, I., Vondran, F. W. R., Le Grand, R., Grimm, D. and Urban, S. (2017). Sodium taurocholate cotransporting polypeptide is the limiting host factor of hepatitis B virus infection in macaque and pig hepatocytes. Hepatology 66(3): 703-716.
- Levrero, M., Testoni, B. and Zoulim, F. (2016). HBV cure: why, how, when? Curr Opin Virol 18: 135-143.
- Li, X., Zhao, J., Yuan, Q. and Xia, N. (2017). Detection of HBV Covalently Closed Circular DNA. Viruses 9(6): E139.
- Liang, T. J., Block, T. M., McMahon, B. J., Ghany, M. G., Urban, S., Guo, J. T., Locarnini, S., Zoulim, F., Chang, K. M. and Lok, A. S. (2015). Present and future therapies of hepatitis B: From discovery to cure. Hepatology 62(6): 1893-1908.
- Long, Q., Yan, R., Hu, J., Cai, D., Mitra, B., Kim, E. S., Marchetti, A., Zhang, H., Wang, S., Liu, Y., Huang, A. and Guo, H. (2017). The role of host DNA ligases in hepadnavirus covalently closed circular DNA formation. PLoS Pathog 13(12): e1006784.
- Margeridon, S., Carrouee-Durantel, S., Chemin, I., Barraud, L., Zoulim, F., Trepo, C. and Kay, A. (2008). Rolling circle amplification, a powerful tool for genetic and functional studies of complete hepatitis B virus genomes from low-level infections and for directly probing covalently closed circular DNA. Antimicrob Agents Chemother 52(9): 3068-3073.
- Mu, D., Yan, L., Tang, H. and Liao, Y. (2015). A sensitive and accurate quantification method for the detection of hepatitis B virus covalently closed circular DNA by the application of a droplet digital polymerase chain reaction amplification system. Biotechnol Lett 37(10): 2063-2073.
- Nassal, M. (2015). HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut 64(12): 1972-1984.
- Ni, Y. and Urban, S. (2017). Hepatitis B Virus infection of HepaRG cells, HepaRG-hNTCP cells, and Primary Human Hepatocytes. Methods Mol Biol 1540:15-25.
- Qu, B., Ni, Y., Lempp, F. A., Vondran, F. W. R. and Urban, S. (2018). T5 exonuclease hydrolysis of hepatitis B virus replicative intermediates allows reliable quantification and fast drug efficacy testing of covalently closed circular DNA by PCR. J Virol 92(23).
- Schreiner, S. and Nassal, M. (2017). A Role for the Host DNA Damage Response in Hepatitis B Virus cccDNA Formation-and Beyond? Viruses 9(5): E125.
- Tu, T., Budzinska, M. A., Shackel, N. A. and Urban, S. (2017). HBV DNA Integration: Molecular mechanisms and clinical implications. Viruses 9(4): E75.
- Xu, C. H., Li, Z. S., Dai, J. Y., Zhu, H. Y., Yu, J. W. and Lu, S. L. (2011). Nested real-time quantitative polymerase chain reaction assay for detection of hepatitis B virus covalently closed circular DNA. Chin Med J (Engl) 124(10): 1513-1516.
- Yu, M., Yang, H., Xie, X., Barauskas, O., Feng, J., Gamelin, L., Xu, S., Delaney, W. and Cheng, G. (2015). 2015 International HBV Meeting. Poster number 43. October 4, 2015-October 8, 2015, Bad Nauheim, Germany.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Qu, B. and Urban, S. (2019). Quantification of Hepatitis B Virus Covalently Closed Circular DNA in Infected Cell Culture Models by Quantitative PCR. Bio-protocol 9(7): e3202. DOI: 10.21769/BioProtoc.3202.
Category
Microbiology > Microbial genetics > Gene expression
Microbiology > Microbial biochemistry > DNA
Molecular Biology > DNA > PCR
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











