- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Extraction of RNA from Recalcitrant Tree Species Paulownia elongata
Published: Vol 8, Iss 14, Jul 20, 2018 DOI: 10.21769/BioProtoc.2925 Views: 7716
Reviewed by: Samik BhattacharyaLaia ArmengotAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation of Nuclei in Tagged Cell Types (INTACT), RNA Extraction and Ribosomal RNA Degradation to Prepare Material for RNA-Seq
Mauricio A. Reynoso [...] Kaisa Kajala
Apr 5, 2018 16205 Views

RNA Purification from the Unicellular Green Alga, Chromochloris zofingiensis
Sean D. Gallaher and Melissa S. Roth
Apr 5, 2018 8150 Views

Simple Method for Efficient RNA Extraction From Arabidopsis Embryos
Fernanda Marchetti [...] Eduardo Zabaleta
Feb 20, 2025 1913 Views
Abstract
Isolation of pure RNA is the basic requisite for most molecular biology work. Plants contain polyphenols and polysaccharides, which can interfere with isolation of pure RNA from them. Especially hardwood tree species like Paulownia elongata have surplus amount of RNA-binding alkaloids, proteins and secondary metabolites that can further complicate the process of RNA extraction. Paulownia elongata is a fast-growing tree species which is known for its role in environmental adaptability and biofuel research. Here we describe an economical, efficient and time-saving method (2 h) to extract RNA from leaf tissues of the tree Paulownia elongata. Lack of DNA contamination and good RNA integrity were confirmed using RNA Gel electrophoresis. The purity of RNA was confirmed using Nanodrop spectrophotometer that revealed an A260:A280 ratio of about 2.0. The purified RNA was successfully used in the downstream applications such as RT-PCR (Reverse Transcription PCR) and qPCR (quantitative PCR). This method could be used for RNA extraction from several other recalcitrant tree species.
Keywords: RNABackground
Paulownia elongata is a widely distributed tree that belongs to the family of Paulowniaceae (Zhu et al., 1986). It is known for its high adaptability and rapid growth rate (Chaires et al., 2017). Paulownia woods are gaining demands from all over the world due to their high stability, low thermal conductivity, decay and rot resistance etc. (Ayrilmis and Kaymakci, 2013). Apart from this, P. elongata is also known to show tolerance to a variety of biotic and abiotic stresses (Chaires et al., 2017). However not many studies are done on understanding its stress tolerance mechanism, which requires extraction of high-quality RNA.
High-quality RNA refers to RNA that is devoid of any genomic DNA, phenols, polysaccharides, secondary metabolites, etc. that interfere with molecular biology techniques (Ouyang et al., 2014). Genomic DNA contamination will affect the detection and quantification of gene expression analysis such as RT-PCR, qPCR, northern blotting and RNA sequencing (Añez-Lingerfelt et al., 2009). This is because the reaction cannot differentiate between cDNA (complementary DNA) and genomic DNA, which will lead to overestimation of gene expression present (Añez-Lingerfelt et al., 2009). Phenols in RNA can oxidize to form quinones that will bind to nucleic acids irreversibly (Ouyang et al., 2014). Polysaccharides and secondary metabolites can co-precipitate and degrade the RNA in the sample thereby affecting the yield, quality and downstream applications of RNA (Ouyang et al., 2014; Ghawana et al., 2011). Since P. elongata leaves are known to have a high content of alkaloids and proteins (Kirov et al., 2014), isolation of pure RNA from them poses a challenge. Methods using spin columns did not yield a good amount of RNA (data not shown) from P. elongata leaves, and this could be due to the fact that spin columns efficiency decreases in the presence of alkaloids (Ouyang et al., 2014). CTAB (cetyltrimethyl ammonium bromide) based methods are spin-column free but are time-consuming (Ouyang et al., 2014). Thus, we combined the Trizol (InvitrogenTM) extraction method and spin-column based purification method in our protocol.
In our study we extracted high quantity RNA using a modified Trizol (InvitrogenTM) method. The quality of RNA was improved by using RNA Clean and Concentrator Kit (ZYMO RESEARCH, USA) with modifications. The RNA yield was measured using Nanodrop spectrophotometer (Figure 1). The Nanodrop measurement peaks can also analyze the presence of phenols or polysaccharides in the sample. In our study, the peak indicated that the RNA was free from phenol or polysaccharide contamination (Figure 1). The integrity of the RNA was further affirmed by running an RNA gel (Formaldehyde free RNA gel kit, Amresco, USA). The gel revealed that the RNA extracted using our method was free from genomic DNA contamination. Clear 28S and 18S rRNA bands were observed (Figure 2). The RNA was then successfully used in downstream applications like RT-PCR and qPCR which amplified the ubiquitin gene (Figures 3 and 4). In RT-PCR, we used cDNA synthesized from our P. elongata RNA as the template. The cDNA was amplified using ubiquitin qPCR primers. The ubiquitin primers used in the study are as follows:
Forward Primer- 5’ GTC AGG AGG AAC ACC TTC TTT 3’
Reverse Primer- 5’ CCT TGA CTG GGA AGA CCA TTAC 3’
Thus bands of about 250 bp were observed as a result of successful amplification of ubiquitin (Figure 3). Our method of RNA isolation not only has high and pure yield but is also time-saving. The entire procedure takes about 2 h.
Materials and Reagents
- Nuclease-free microfuge tubes
- Nuclease-free micropipette tips
- Nitrile powder free gloves
- Liquid nitrogen
- RNase (ribonuclease) Away (Thermo Fisher Scientific, catalog number: 7000TS1 )
- Trizol (Thermo Fisher Scientific, InvitrogenTM, catalog number: 15596026 )
- 24:1 Chloroform:Isoamyl alcohol (Acros Organics, catalog number: 327155000 )
- 75% and 100% ethanol
- Nuclease-free water (Not DEPC-treated) (Thermo Fisher Scientific, InvitrogenTM, catalog number: 4387936 )
- Turbo DNase (deoxyribonuclease) (Thermo Fisher Scientific, AmbionTM, catalog number: AM2238 )
- RNA Clean and Concentrator kit (ZYMO RESEARCH, catalog number: R1015 )
- Chilled isopropanol
Equipment
- Pipettes (BioExpress, GeneMate, catalog number: P-3960-20 )
- Mortar and pestle (Harold Import, HIC, catalog number: 43717 )
- Vortexer (BioExpress, GeneMate, catalog number: S-3200-1 )
- Refrigerated microcentrifuge (Labnet International, catalog number: C2500-R )
- Water bath
- -80 °C freezer
- Fume hood (KEWAUNEE, model: H05_5460-00 )
- Nanodrop Spectrophotometer (Thermo Fisher Scientific, USA)
Procedure
- RNA Extraction
Note: It is modified from standard TrizolTM procedure.- Spray gloves, equipment and all materials using RNase (ribonuclease) AWAY.
- Snap freeze about 100 mg of leaves by immersing the leaves for 5 sec in liquid nitrogen.
Tip: Snap freezing works better than crushing the leaves directly in liquid nitrogen. - Grind the frozen leaves in the presence of 1 ml of Trizol using a mortar and pestle (not necessary to be cold) and transfer to a nuclease-free microfuge tube using a pipette.
Note: This step should be performed inside a fume hood. - Incubate the tube for 5 min at room temperature (RT).
- Then add 200 μl of 24:1 Chloroform:Isoamyl alcohol into the tube and vortex to mix thoroughly.
Tip: The sample must be vortexed until it is homogeneously green. This vortexing step is critical for high yield of RNA. - Incubate this mixture for 2 min at RT.
- Then centrifuge at 12,000 x g for 15 min at 4 °C.
- Transfer the supernatant (the aqueous phase containing RNA) to a new tube.
Tip: Note the volume of phase transferred. - Add chilled isopropanol into the new tube at a volume of 0.5 times the original volume of aqueous phase present.
Tip: Isopropanol must be chilled, otherwise the yield of RNA might be low. - Vortex the mixture vigorously for at least 10 sec and incubate for about 15 min at -20 °C.
- Centrifuge the mix for 10 min at 12,000 x g at 4 °C.
- RNA can be seen as a small white pellet at the bottom of the tube.
- Remove the supernatant isopropanol without disturbing the pellet. Wash the RNA pellet by adding 500 μl of 75-80% ethanol and discard the supernatant after spinning at 7,000 x g for 5 min (Potential stop point: can be stored at -20 °C for up to a week).
- Spin the tube again (without adding any reagents) and then remove any residual ethanol that sticks to the wall of the microcentrifuge tube.
- Air dry the pellet for 15 min by placing the inverted tube inside the fume hood.
- Then resuspend the pellet in 40 μl of nuclease-free water and incubate in a water bath with the tube open at 60 °C for 15 min to completely dissolve it. The cap of microfuge tube should be kept open for efficient evaporation of residual ethanol (Potential Stop point: samples can be stored at -80 °C). The samples stored at -80 °C must be thawed on ice and treated by the following steps.
- Spray gloves, equipment and all materials using RNase (ribonuclease) AWAY.
- RNA Purification
Note: It is modified from ZYMO Clean and Concentrator KitTM protocol (ZYMO RESEARCH, USA).- Add 80 μl of RNA binding buffer to the RNA (see above) and gently mix it using pipette tip.
- Then add 120 μl of 100% ethanol to the tube followed by gentle mixing.
- Transfer the RNA sample to a Zymospin column in a collection tube and spin at 12,000 x g for 30 sec at 4 °C.
- Wash the column with 400 μl of RNA wash buffer by spinning at 12,000 x g for 30 sec at 4 °C. The flow-through will be discarded.
- Prepare DNase cocktail by mixing 5 μl of Turbo DNase (Thermo Fisher Scientific, AmbionTM) and 35 μl of DNA digestion buffer (ZYMO RESEARCH, USA) gently in a microfuge tube.
- Add the cocktail to the RNA column and incubate for 20 min at RT.
Tip: Longer incubation times do not improve purity. - Add about 400 μl of RNA Prep buffer (or RNA Pre-Wash Buffer) to the column and spin at 12,000 x g for 30 sec at 4 °C. Then discard the flow through.
- Wash the column again twice using 400 μl of RNA Wash buffer like Step B4.
- Spin the column (without adding any reagents) for 2 min to remove the residual ethanol.
- Finally, transfer the column to a new nuclease-free microfuge tube.
- Add about 20 μl of nuclease-free water to the column and incubate for 2 min at RT. Collect the flow through after spinning for 2 min at 12,000 x g at 4 °C.
- Pipette out the flow through and add it to the column again.
- Incubate the column for 2 min at RT and collect the pure RNA after spinning for 2 min at 12,000 x g at 4 °C.
Data analysis
- RNA measurement: The concentration of extracted RNA was measured using a Nanodrop spectrophotometer. Four biological replicates were used. The spectrophotometric images (Figure 1) showed that the RNA we extracted was of high yield starting from 240 ng. Pure RNA is considered to have an A260:A280 ratio between 1.8 and 2.2 (Ouyang et al., 2014). Since our RNA had ratios of 2.06-2.11, it revealed that this extraction protocol yields pure RNA. The single clear peak at 260 nm shows that the RNA extraction is devoid of any contaminants.

Figure 1. Nanodrop spectrophotometer images of RNA extracted from P. elongata using the modified method. The 260/280 ratios are 2.09 (A), 2.07 (B), 2.06 (C) and 2.11 (D). Four pictures represent four different biological samples.
RNA integrity: The RNA integrity was assessed by running an RNA gel. The RNA samples extracted without following our purification procedure was compared to RNA samples extracted following our modified protocol. About 5 μl of each RNA sample was loaded onto each well. The RNA gel revealed clear, distinct 18S and 28S ribosomal RNA bands for samples obtained using our protocol (Figure 2). The RNA samples obtained with other methods had smears indicating that there is genomic DNA contamination.
Figure 2. RNA gel pictures. A1, A2, A3: P. elongata RNA without our purification procedure. B1, B2, B3, B4: P. elongata RNA extracted using modified method 1-4 indicates the different biological controls (The picture’s color has been altered to grayscale).
RNA downstream applications: To test the efficiency of the extracted RNA, the samples were used in RT-PCR and qPCR as templates. PCR products were successfully amplified using ubiquitin primers from the cDNA reverse-transcribed from the extracted RNA. The PCR conditions used were–initial denaturation at 95 °C for 5 min, 30 PCR cycles with denaturation at 95 °C for 30 sec, annealing at 50 °C for 30 sec and extension at 72 °C for 45 sec, with a final extension at 72 °C for 7 min. Four biological replicates were used for RT-PCR. Clear bands of ubiquitin were observed at about 250 bp (Figure 3). The integrity of mRNA (messenger RNA) was tested using qPCR. The qPCR conditions were: initial denaturation at 95 °C for 30 sec, 39 cycles with denaturation at 95 °C for 5 sec, annealing at 50 °C for 30 sec followed by melting-curve analysis. Four biological replicates and 3 technical replicates were used for qPCR.
The amplification curve obtained from qPCR (Figure 4) showed that the mRNA was not degraded in the total RNA sample and that the ubiquitin gene was present in all 4 biological samples. The melt curve picture reveals that the ubiquitin primers were highly specific and that the results are not due to false positives (Figure 4). These results showed that the quality of RNA we extracted is suitable for basic experiments like PCR and qPCR.
Figure 3. RT-PCR gel for ubiquitin gene using RNA extracted by this method (Ubiquitin bands of 250 bp size are observed in all four lanes from 3-6). Lane 1: Ladder; Lane 2: Negative control (water); Lanes 3-6: P. elongata cDNA amplified by ubiquitin primers using RNA extracted from 4 different biological controls.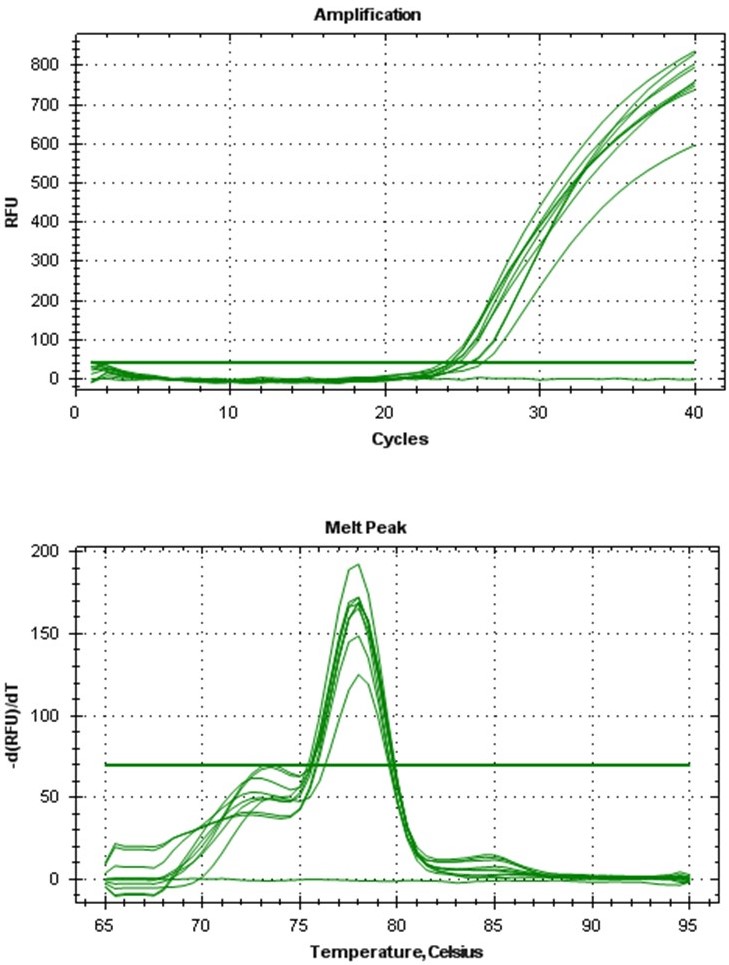
Figure 4. Amplification curve (top) and melt curve (bottom) observed in qPCR of ubiquitin gene from P. elongata cDNA - Conclusion
In our study, we developed a protocol for efficient isolation of high-quality RNA from the leaves of a recalcitrant tree species P. elongata. Traditional methods that use CTAB or spin columns were not able to produce this quality of RNA with high yield (data not shown), due to the presence of high amount of alkaloids and proteins in the leaves of P. elongata. The method we developed is rapid (2 h), universal and high yielding. We believe that this method could be used for RNA isolation from various recalcitrant plant species.
Acknowledgments
The authors declare no conflict of interests. The authors want to acknowledge the following two awards/scholarships awarded to NR at the California State University, Northridge: Thesis/Project /Dissertation/Support Award and Richard Duenckel Gardens Club Scholarship. This protocol has been used in our recently published research paper (Ramadoss et al., 2018).
References
- Añez-Lingerfelt, M., Fox, G. E. and Willson, R. C. (2009). Reduction of DNA contamination in RNA samples for reverse transcription-polymerase chain reaction using selective precipitation by compaction agents. Anal Biochem 384(1): 79-85.
- Ayrilmis, N. and Kaymakci, A. (2013). Fast growing biomass as reinforcing filler in thermoplastic composites: Paulownia elongata wood. Ind Crops Prod 43: 457-464.
- Chaires, M., Gupta, D., Joshee, N., Cooper, K. K. and Basu C. (2017) RNA-seq analysis of the salt stress-induced transcripts in fast-growing bioenergy tree, Paulownia elongata. Journal of Plant Interactions. 12(1):128-136.
- Ghawana, S., Paul, A., Kumar, H., Kumar, A., Singh, H., Bhardwaj, P. K., Rani, A., Singh, R. S., Raizada, J., Singh, K. and Kumar, S. (2011). An RNA isolation system for plant tissues rich in secondary metabolites. BMC Res Notes 4: 85.
- Kirov, V., Shindarska, Z., Kostadinova, G., Gencheva, A., Hadgiev, S., Penev, T. and Baykov, B. (2014). Comparative study of new energy crops for the production of biogas. Int J Curr Microbiol App Sci 3(11): 181-188.
- Ouyang, K., Li, J., Huang, H., Que, Q., Li, P. and Chen, X. (2014). A simple method for RNA isolation from various tissues of the tree Neolamarckia cadamba. Biotechnol Biotechnol Equip 28(6): 1008-1013.
- Ramadoss, N., Gupta, D., Vaidya, B. N., Joshee, N. and Basu, C. (2018). Functional characterization of 1-aminocyclopropane-1-carboxylic acid oxidase gene in Arabidopsis thaliana and its potential in providing flood tolerance. Biochem Biophys Res Commun. doi: 10.1016/j.bbrc.2018.06.036.
- Zhu, Z. H., Chao, C. J., Lu, X. Y. and Xiong Y. G. (1986). Paulownia in China: cultivation and utilization by Chinese academy of forestry staff. Asian Network for Biological Sciences and International Development Research Centre.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Ramadoss, N. and Basu, C. (2018). Extraction of RNA from Recalcitrant Tree Species Paulownia elongata. Bio-protocol 8(14): e2925. DOI: 10.21769/BioProtoc.2925.
Category
Plant Science > Plant molecular biology > RNA > RNA extraction
Molecular Biology > RNA > RNA extraction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










