- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Measurement of Oxygen Consumption Rate (OCR) and Extracellular Acidification Rate (ECAR) in Culture Cells for Assessment of the Energy Metabolism
Published: Vol 8, Iss 10, May 20, 2018 DOI: 10.21769/BioProtoc.2850 Views: 76078
Reviewed by: Amriti Rajender LullaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Assessment of Cellular Redox State Using NAD(P)H Fluorescence Intensity and Lifetime
Thomas S. Blacker [...] Gyorgy Szabadkai
Jan 20, 2017 14865 Views

FACS-based Glucose Uptake Assay of Mouse Embryonic Fibroblasts and Breast Cancer Cells Using 2-NBDG Probe
Shengli Dong and Suresh K Alahari
Apr 20, 2018 11540 Views

Optimization of Extracellular Flux Assay to Measure Respiration of Anchorage-independent Tumor Cell Spheroids
Zaineb Javed [...] Nadine Hempel
Feb 20, 2022 4575 Views
Abstract
Mammalian cells generate ATP by mitochondrial (oxidative phosphorylation) and non-mitochondrial (glycolysis) metabolism. Cancer cells are known to reprogram their metabolism using different strategies to meet energetic and anabolic needs (Koppenol et al., 2011; Zheng, 2012). Additionally, each cancer tissue has its own individual metabolic features. Mitochondria not only play a key role in energy metabolism but also in cell cycle regulation of cells. Therefore, mitochondria have emerged as a potential target for anticancer therapy since they are structurally and functionally different from their non-cancerous counterparts (D'Souza et al., 2011). We detail a protocol for measurement of oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) measurements in living cells, utilizing the Seahorse XF24 Extracellular Flux Analyzer (Figure 1). The Seahorse XF24 Extracellular Flux Analyzer continuously measures oxygen concentration and proton flux in the cell supernatant over time (Wu et al., 2007). These measurements are converted in OCR and ECAR values and enable a direct quantification of mitochondrial respiration and glycolysis. With this protocol, we sought to assess basal mitochondrial function and mitochondrial stress of three different cancer cell lines in response to the cytotoxic test lead compound mensacarcin in order to investigate its mechanism of action. Cells were plated in XF24 cell culture plates and maintained for 24 h. Prior to analysis, the culture media was replaced with unbuffered DMEM pH 7.4 and cells were then allowed to equilibrate in a non-CO2 incubator immediately before metabolic flux analysis using the Seahorse XF to allow for precise measurements of Milli-pH unit changes. OCR and ECAR were measured under basal conditions and after injection of compounds through drug injection ports. With the described protocol we assess the basic energy metabolism profiles of the three cell lines as well as key parameters of mitochondrial function in response to our test compound and by sequential addition of mitochondria perturbing agents oligomycin, FCCP and rotenone/antimycin A.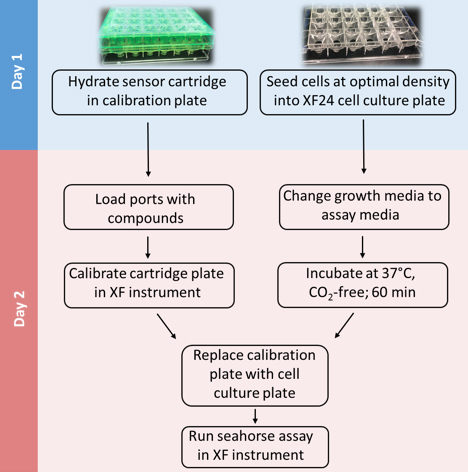
Figure 1. Overview of seahorse experiment
Background
Natural products are small molecules that are isolated from natural sources. Over the last century, these molecules have been instrumental in treating human diseases, especially inspired chemotherapeutics. Metabolites like taxol, vincristine, and doxorubicin have revolutionized how we treat malign cancers and other natural products, for example rapamycin, oligomycin, and bafilomycin, are used as molecular probes and enable molecular studies of biochemical and cellular processes in the laboratory. While studying the mechanism of action of the cytotoxic natural product mensacarcin, we found that a fluorescently labeled mensacarcin probe localizes to a great extent in mitochondria (Plitzko et al., 2017). To investigate if mensacarcin’s cytotoxic properties might be derived from interference with mitochondrial function, we sought to examine mensacarcin’s effects on cellular bioenergetics. Using a Seahorse Extracellular Flux Analyzer, we monitored cellular oxygen consumption rates (OCR) and extracellular acidification rates (ECAR) in real time as measures of mitochondrial respiration and glycolysis, respectively (Wu et al., 2007; Serill et al., 2015). The Seahorse XF24 Extracellular Flux Analyzer allows continuous direct quantification of mitochondrial respiration and glycolysis of living cells. The instrument uses a sensor cartridge in a 24-well plate format with each sensor being equipped with two embedded fluorophores: one which is quenched by oxygen (O2) and the other that is sensitive to change in pH. During measurements, the sensor cartridge is lowered 200 µm above the cell monolayer, forming a micro-chamber of about 2 µl. The Seahorse instrument contains fiber optic bundles that emit light, excite the fluorophores, and then measures the change in the fluorophore’s emission. The very small test volume formed by the transient micro chamber allows for sensitive, precise, and nondestructive measurements of parameters in real time. Changes in oxygen concentration and pH are automatically calculated and reported as Oxygen Consumption Rate (OCR) and Extra Cellular Acidification Rate (ECAR). Once a measurement is completed, the sensors lift which allows the larger medium volume above to mix with the medium in the transient micro chamber, restoring values to baseline. The sensor cartridge contains ports that allow sequential addition of up to four compounds per well during the assay measurements.
With the described protocol we assessed the energy metabolism of three cell lines (HCT-116, SK-Mel-28, and SK-Mel-5) (Figure 6). Addition of mensacarcin was found to have pronounced effect on the basal OCR of melanoma cells and no increasing effect on ECAR. An increase in glycolysis is often observed as a compensatory response. Mitochondria are essential for the energy metabolism of cells and have a key role in apoptotic cell death. Alteration of the mitochondrial respiration or the equilibrium between the pro-apoptotic and anti-apoptotic proteins can induce mitochondrial failure. To gain insights into the induced mitochondrial impairment in melanoma cells, we assessed key parameters of mitochondrial respiration by consecutively exposing cells to well described mitochondria perturbing reagents. Following addition of our test compound mensacarcin, we sequentially added oligomycin, FCCP, and lastly rotenone and antimycin A (Figure 5). Oligomycin inhibits ATP synthase and reduces OCR, FCCP uncouples oxygen consumption from ATP production and raises OCR to a maximal value, and antimycin A and rotenone target the electron transport chain and reduce OCR to a minimal value. The mitochondria stress test protocol provides information on basal respiration, ATP-linked respiration, proton leak, maximal respiration capacity, and non-mitochondrial respiration of cells. Therefore, this assay can be used to provide insight on the mechanism of action of compounds that directly target mitochondrial respiration.
Traditional measurements of mitochondrial function or glycolysis require an oxygen electrode, or kits and dyes that utilize colorimetric or fluorimetric detection (Li and Graham, 2012; TeSlaa and Teitell, 2014). Most of these methods are invasive and cumbersome methods that only allow low sample throughput. In contrast, the Seahorse analyzer assay with its sensor cartridge system enables measurement of mitochondrial respiration and glycolysis in real time and in a non-invasive manner that does not require any dyes or labels. Cellular energy metabolism research is highly topical in all fields of mammalian cell biology. The following protocol was developed for researchers in cancer biology but with approaches that suit studies of energy metabolism in all mammalian cell systems.
Materials and Reagents
- CELLSTAR® Tissue Culture Plates, 96-well (Greiner Bio One International, catalog number: 655180 )
- Sterile racked pipette tips (1 ml and 200 μl) (VWR, catalog numbers: 613-0738 ; 613-0742 )
- Sterile basins (Corning, Costar®, catalog number: 4870 )
- Sterile reagent tubes (15 and 50 ml) (VWR, catalog numbers: 89039-668 ; 89039-662 )
- Sterile Serological pipettes (5, 10, 25, 50 ml) (Fisher Scientific, catalog numbers: 13-678-11 , 13-678-11D , 13-678-11E , 13-678-11F )
- Glass bottles (500 ml) (Fisher Scientific, catalog number: FB8001000 )
- HCT-116, SK-Mel-5 and SK-Mel-28 cells (ATCC, catalog numbers: CCL-247 , HTB-70 , HTB-72 )
- Seahorse XF24 FluxPak (including sensor cartridges, tissue culture plates, calibrant solution and calibration plates) (Agilent Technologies, Santa Clara, CA)
- Trypsin/EDTA (0.25%/2.21 mM) (Corning, catalog number: 25-053-Cl )
- 1x Ca2+/Mg2+-free DPBS (Thermo Fisher Scientific, GibcoTM, catalog number: 14190250 )
- Liquid Dulbecco’s modified Eagle’s medium (DMEM) (Corning, catalog number: 10-013 )
- Fetal bovine serum (FBS) (Atlanta Biologicals, catalog number: S11150 )
- Penicillin/streptomycin solution 100x (Corning, catalog number: 30-002-Cl )
- Powder Dulbecco’s modified Eagle’s medium (DMEM) without Na2HCO3, without HEPES (Corning, catalog number: 50-013 )
- Sodium hydroxide (NaOH) (VWR, catalog number: 97064-476 )
- Oligomycin (Merck, catalog number: 495455-10MG )
- DMSO (VWR, catalog number: BDH1115-1LP )
- FCCP (Cayman Chemical, catalog number: 15218 )
- Rotenone (Cayman Chemical, catalog number: 13995 )
- Antimycin A (Enzo Life Sciences, catalog number: ALX-380-075-M005 )
- Culture media (10% (v/v) FBS) (see Recipes)
- Assay media (see Recipes)
- NaOH (1 M) (see Recipes)
- Oligomycin (10 µM) (see Recipes)
- FCCP (5 µM) (see Recipes)
- Rotenone (5 µM)/antimycin A (5 µM) (see Recipes)
Equipment
- Hemacytometer (Hausser Scientific, catalog number: 1490 )
- Biological Safety Cabinet Class II, Type A2 (NuAire, model: NU-425-400ES )
- Seahorse XF Extracellular Flux Analyzer (Agilent Technologies, Santa Clara, CA)
- Pipet-Lite Pipette XLS STD 20 XLS (Mettler Toledo, Rainin, model: SL-2XLS+ )
- Pipet-Lite Pipette XLS STD 200 (Mettler Toledo, Rainin, model: SL-200XLS+ )
- Pipet-Lite Pipette XLS 1000 (Mettler Toledo, Rainin, model: SL-1000XLS+ )
- Multichannel Pipet-Lite Pipette XLS 8-CH 1200 (Mettler Toledo, Rainin, model: L8-1200XLS+ )
- Multichannel Pipet-Lite Pipette XLS 8-CH 200 (Mettler Toledo, Rainin, model: L8-200XLS+ )
- Aspirator pump
- Humidified non-CO2 incubator (XF Prep Station; Agilent Technologies, Santa Clara, CA)
- Shallow water bath (Thermo Fisher Scientific, Thermo ScientificTM, model: Precision 180 )
- Pipette controller (BrandTech Scientific, model: Accu-Jet® Pro , catalog number: 26330)
- Humidified, 37 °C, 5% CO2 incubator (Eppendorf, model: Galaxy® 170 R )
- -20 °C biomedical freezer (Sanyo, model: MDF-U731M )
- Autoclave (Consolidated Sterilizer Systems, model: SSR-3A , ADVPB)
- Inverted light microscope (Nikon Instruments, model: Eclipse TS100 )
- pH-meter with semi-micro electrode (Thermo Fisher Scientific, Thermo ScientificTM, model: Orion StarTM A211 , with ROSS 8103BN electrode: (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 8103BN )
Software
- Seahorse Bioscience XF24 software
- Excel (Microsoft)
- GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA)
Procedure
- Optimization of seeding density
In an initial experiment, the optimal seeding density is required for each cell type. Typically, the cell density ranges from 10,000 to 60,000 cells per well and can vary widely among cell lines. A first point of orientation can be the cell number that gives confluency of approx. 95% overnight in a 96-well cell culture plate as the seeding surface is comparable to the seahorse culture plate. The seeding number should give a confluent and healthy and consistent monolayer on the day of the assay.- HCT-116, SK-Mel-5 and SK-Mel-28 cells were seeded in a Seahorse XF24 cell culture plate at various concentrations ranging from 10,000 to 30,000 cells/well with a two-step seeding technique as described below in Procedure B (Figure 2). Seeding cells in triplicates is recommended.
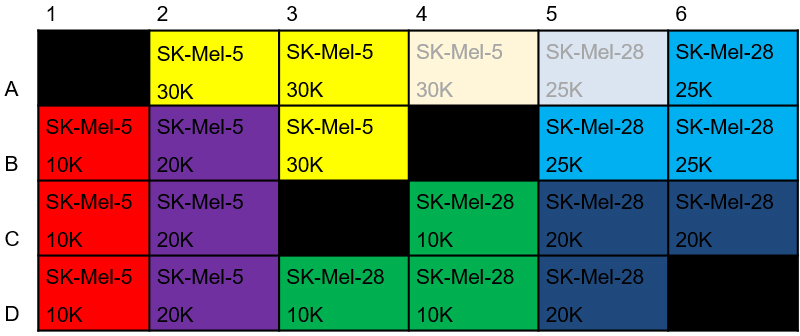
Figure 2. Plate layout for cell density evaluation. Shown here is the exemplary seeding layout for the SK-Mel-5 and SK-Mel-28 cell lines (seeding density for HCT-116 cells was evaluated on a second plate; not shown). - Cells were then assayed in the XF24 instrument as described in Procedure E (without loading compounds into ports) using Table 1 commands.
Table 1. Protocol commands for cell density evaluation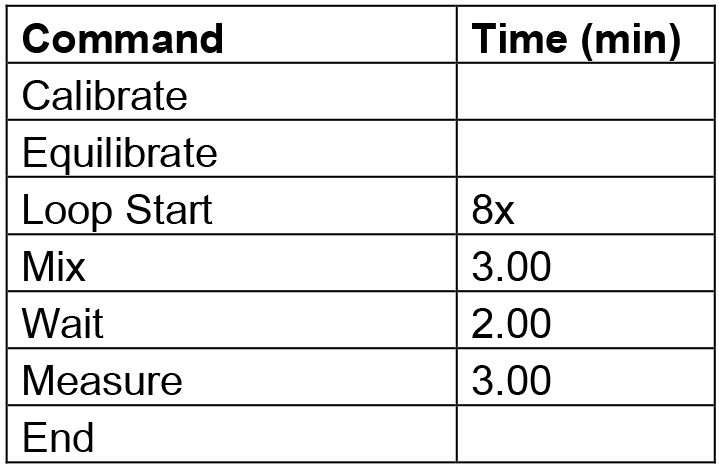
As seen in Figure 3, a linear increase of OCR values with increasing cell density was observed in all three cell lines. ECAR values begin to level off at 20,000 cells/well for SK-Mel-28 and SK-Mel-5 while being much lower and steadily increasing for HCT-116. Thus, a seeding number of 20,000 cells/well for SK-Mel-28 and SK-Mel-5 and of 35,000 cells/well for HCT-116 were chosen to ensure being within the linear response range while having high reading values to observe increases as well as decreases in OCR and ECAR.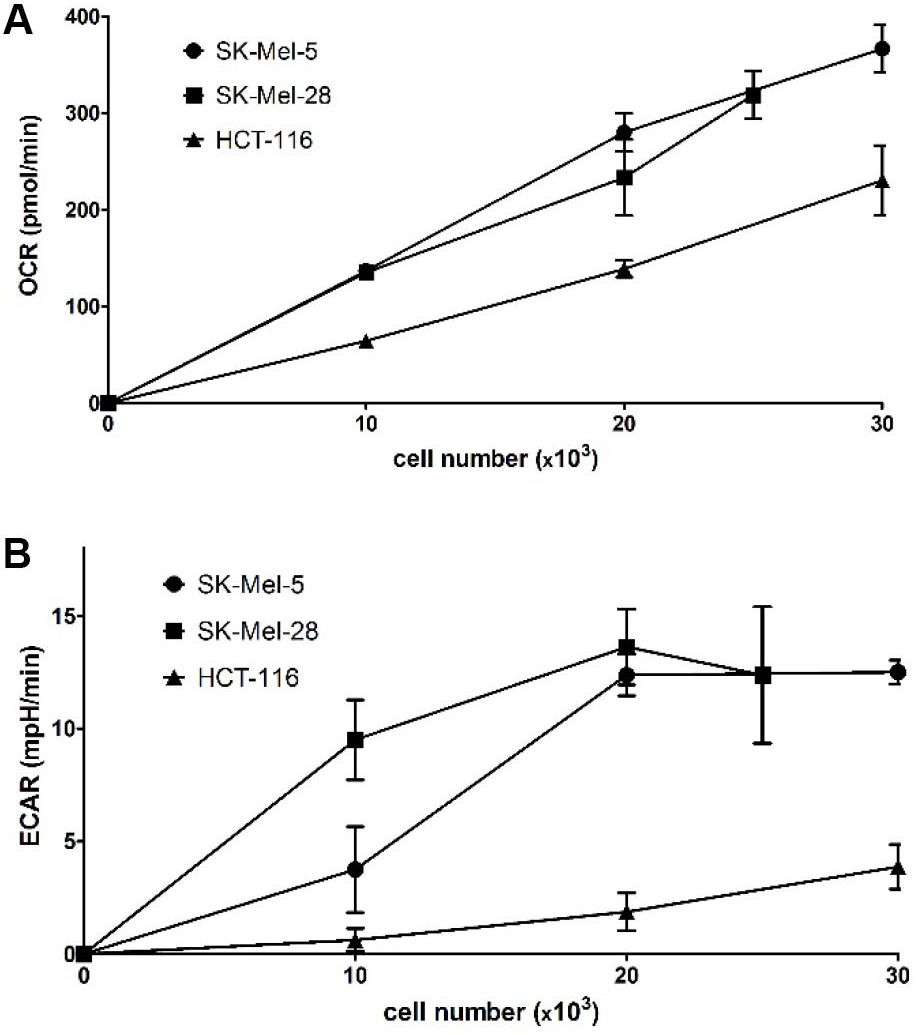
Figure 3. Optimization of assay conditions: evaluation of OCR and ECAR depending on the seeding density of three different cell lines
- HCT-116, SK-Mel-5 and SK-Mel-28 cells were seeded in a Seahorse XF24 cell culture plate at various concentrations ranging from 10,000 to 30,000 cells/well with a two-step seeding technique as described below in Procedure B (Figure 2). Seeding cells in triplicates is recommended.
- Seeding cells into Seahorse XF24 tissue culture plate (Day 1)
Note: The seeding and growing of cells are performed with good sterile cell culture technique. A two-step seeding method is used to obtain a consistent even monolayer which is vital to obtain consistent and accurate data:- Pre-warm culture media, trypsin solution and DPBS to 37 °C.
- For adherent cells, wash cells with DPBS, and add trypsin and wait until cells begin to detach. Add culture media with serum to deactivate trypsin and pipette up and down to create a uniform cell suspension. Count cells with a hemocytometer and resuspend cells in growth media to the desired final concentration to seed in 100 µl.
- Plate 100 µl cell suspension into a Seahorse XF24 tissue culture plate. Put media only (no cells) in the background correction wells (A1, B4, C3, D6).
- Let the culture plate sit for 1 h in the bio-hood without moving it around (in order to let cells settle evenly).
- Place the culture plate into an incubator (37 °C, 5% CO2) for 4 h.
- Carefully add 150 µl growth media (final volume in well 250 µl). Hold the pipette tip at an angle and add to the well side to not destroy even layer of newly attached cells.
- Let cells grow overnight at 37 °C, 5% CO2.
- Pre-warm culture media, trypsin solution and DPBS to 37 °C.
- Hydrate sensors (Day 1)
- Open XF 24 FluxPak and take out the sensor cartridge (green) and calibration plate (clear) (Figure 4).
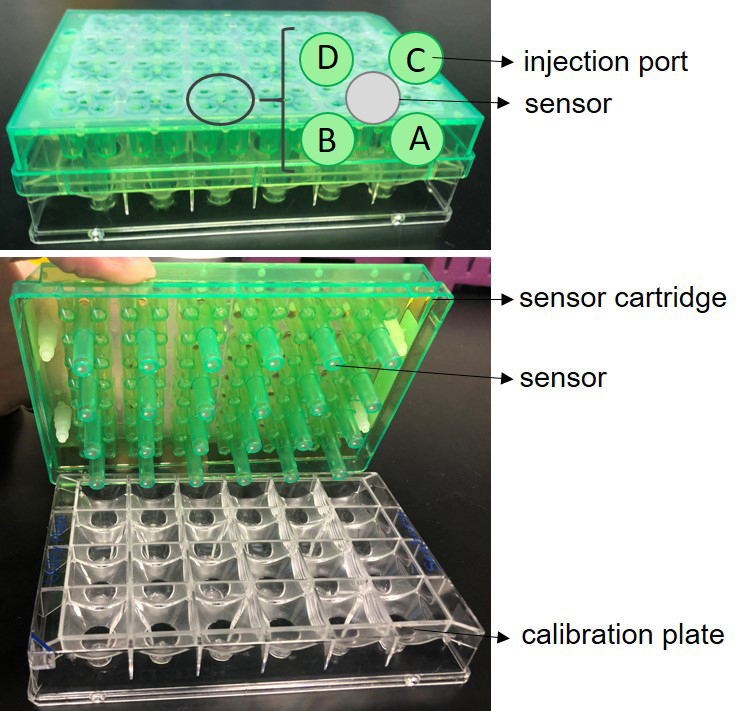
Figure 4. Seahorse XF 24 sensor cartridge. A. The sensor cartridge sitting on top of a calibration plate with injection ports shown. B. Bottom side of the sensor plate which shows sensors with embedded fluorophores. - Place the sensor cartridge (sensors up) next to the calibration plate (be careful not to touch sensors).
- Fill each well of the calibration plate with 1 ml of Seahorse XF Calibrant.
- Lower the sensor cartridge onto the calibrant plate submerging the sensors in calibrant (be careful not to touch walls with sensors).
- Place in a non-CO2 37 °C incubator overnight. To prevent evaporation of the XF Calibrant, verify that the incubator is properly humidified.
- Open XF 24 FluxPak and take out the sensor cartridge (green) and calibration plate (clear) (Figure 4).
- Stabilization of instrument (Day 1)
- Turn on an XF24 Analyzer, open Seahorse Bioscience software and log in.
- Write the assay template. When planning and writing the assay protocol be careful not to create a protocol that is longer than cells can manage without CO2 in unbuffered media. Depending on cell type this is 2-3 h. If in doubt, a cell viability assay can be performed after the seahorse assay.
- Leave the XF24 Analyzer on overnight with XF24 software running and logged in to ensure equilibration to 37 °C.
- Turn on an XF24 Analyzer, open Seahorse Bioscience software and log in.
- Seahorse assay (Day 2)
- Check on the confluency of cells. Evenly spacing of cells is needed, without large cell clumps or blank patches, as this could impair the accuracy of data.
- Pre-warm assay media to 37 °C.
- Pre-warm compounds and adjust to pH 7.4 with NaOH (1 M) if necessary.
- Perform media exchange in a Seahorse XF24 tissue culture plate:
- Remove 150 µl growth media with a multichannel pipet.
- Add 1 ml assay media with a multichannel pipette.
- Remove 1 ml with a multichannel pipette.
- Add 475 µl assay media with a multichannel pipette (575 µl final volume).
- Place the cell plate into a CO2-free incubator for approx. 60 min.
- Remove 150 µl growth media with a multichannel pipet.
- Load cartridge with desired compounds:
- Pre-warm compounds to 37 °C.
- Load 50-100 µl of compound into appropriate port of cartridge (for mitochondrial stress test: 64 µl into port A, 71 µl port B, 79 µl port C, 88 µl port D). (see Note 1) Load equivalent amounts of assay media into equivalent port for background wells (see Note 2).
- Place back into the incubator (non-CO2) for 10 min to allow heating up to 37 °C again. Handle carefully, carry only by holding onto the calibration plate. Move as less as possible.
- Pre-warm compounds to 37 °C.
- Calibration and running seahorse assay:
- Load assay template in Seahorse XF24 software.
- Press green ‘START’ button.
- Make sure to load the correct protocol, the correct save directory and saving name.
- Press ‘START’.
- Load sensor cartridge with calibration plate into instrument tray (the notch goes in the front, left corner. Make sure that the plate sits correctly and flat, between all 8 tabs)
- Follow the instructions on the screen in order to calibrate and equilibrate sensors.
- Once equilibration step is done, remove the calibration plate and replace with cell culture plate.
- Load assay template in Seahorse XF24 software.
- Check on the confluency of cells. Evenly spacing of cells is needed, without large cell clumps or blank patches, as this could impair the accuracy of data.
- Protocol commands (mitochondria stress test, Table 2, Figure 5)
Table 2. Protocol commands for mitochondrial stress test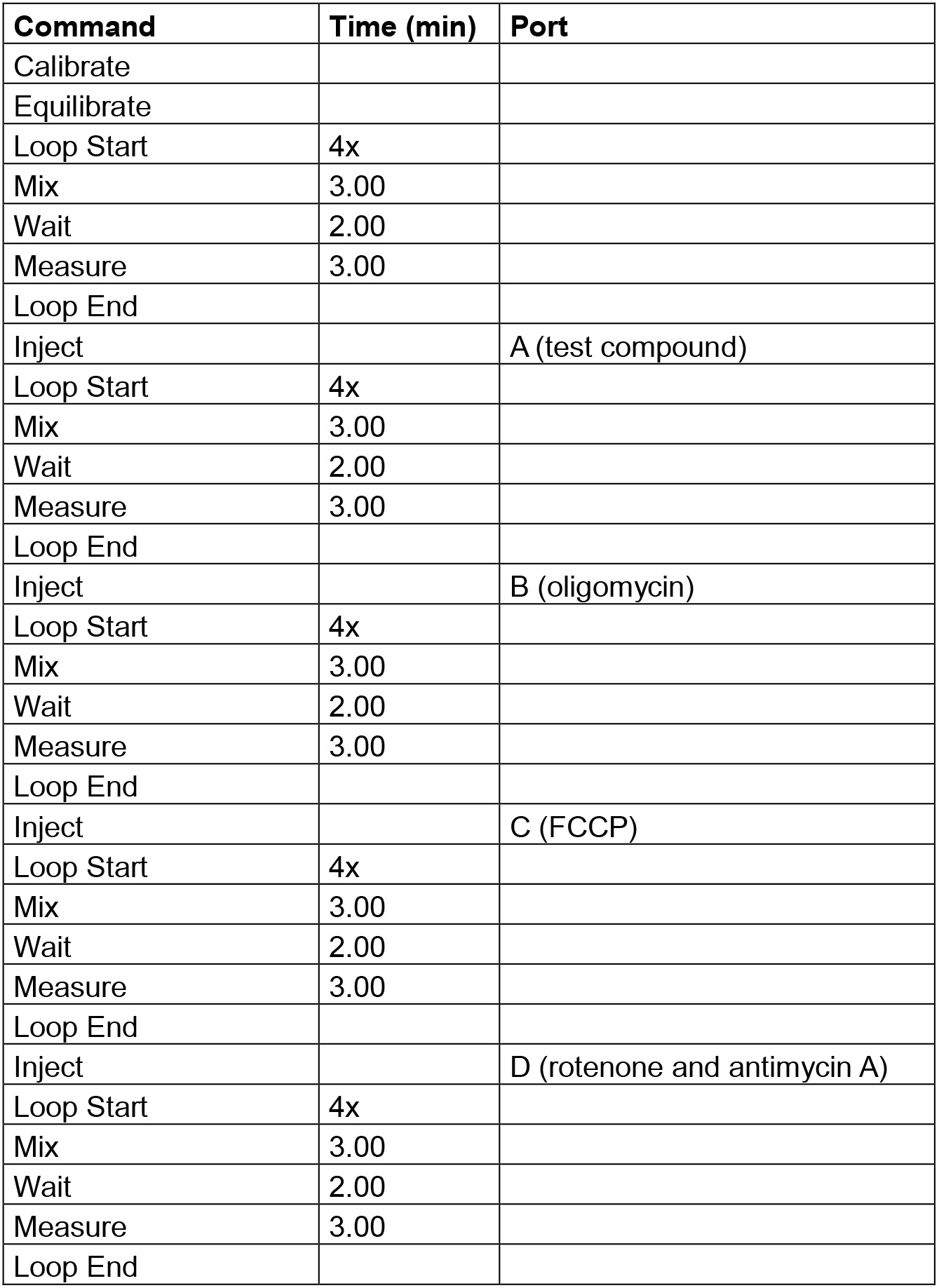
- Pipet into ports with angle, do not touch the bottom, do not tap to prevent leakage. The liquid is only held by capillary forces.
- It is mandatory to load ports for the background wells with assay media that contains the same concentration of DMSO as the compounds to account for any DMSO effects on cells.
- Once injected into the wells, compounds are diluted 1:10. This will give a final concentration of 1 µM oligomycin and 0.5 µM FCCP, rotenone and antimycin A, respectively, in the cell culture well.
- Culture media (10% (v/v) FBS)
Note: Work under sterile conditions in a laminar flow hood.- Open liquid DMEM bottle
- Take out 55 ml with a sterile Serological pipette and discard the liquid
- Add 50 ml FBS with a sterile Serological pipette
- Add 5 ml penicillin/streptomycin solution
- Store at 4 °C
- Open liquid DMEM bottle
- Assay media (sterile, unbuffered, 250 ml)
Note: Work under sterile conditions in a laminar flow hood.- Autoclave 250 ml ultrapure H2O in a glass bottle
- Dissolve 3,34 g powder DMEM without NaHCO3 and without HEPES in 250 ml autoclaved H2O
- Warm to 37 °C
- Adjust to pH 7.40 with NaOH (1 M)
- Store at 4 °C
- Autoclave 250 ml ultrapure H2O in a glass bottle
- NaOH (1 M)
Dissolve 4 g NaOH pellets in 100 ml autoclaved H2O - Oligomycin (10 µM)
- Prepare freshly on the day of seahorse assay (day 2) (see Note 1)
- Prepare 1 mM solution in 1 ml DMSO: Dissolve 0.7911 mg oligomycin in DMSO
- Dilute to 10 µM in assay media (1% DMSO): Pipet 20 µl of 1 mM oligomycin into 1,980 µl assay media
- Warm to 37 °C and adjust to pH 7.4 with NaOH (1 M) if necessary
- Prepare freshly on the day of seahorse assay (day 2) (see Note 1)
- FCCP (5 µM)
- Prepare freshly on the day of seahorse assay (day 2) (see Note 1)
- Prepare 50 mM solution in DMSO: Dissolve 2.54 mg FCCP in 200 µl DMSO
- Dilute to 500 µM: Pipet 10 µl of 50 mM FCCP into 990 µl DMSO
- Dilute to 5 µM in assay media (1% DMSO): Pipet 20 µl of 500 µM FCCP into 1,980 µl assay media
- Warm to 37 °C and adjust to pH 7.4 with NaOH (1 M) if necessary
- Prepare freshly on the day of seahorse assay (day 2) (see Note 1)
- Rotenone (5 µM)/antimycin A (5 µM)
- Prepare freshly on the day of seahorse assay (day 2) (see Note 1)
- Prepare 50 mM solution in DMSO: Solve 3.94 mg rotenone and 5.49 mg antimycin A in 200 µl DMSO
- Dilute to 1 mM: Pipet 10 µl of 50 mM rotenone/antimycin A into 490 µl DMSO
- Dilute to 5 µM in assay media (0.5% DMSO): Pipet 20 µl of 1 mM rotenone/antimycin A into 1,980 µl assay media
- Warm to 37 °C and adjust to pH 7.4 with NaOH (1 M) if necessary
- Prepare freshly on the day of seahorse assay (day 2) (see Note 1)
- D’Souza, G. G., Wagle, M. A., Saxena, V. and Shah, A. (2011). Approaches for targeting mitochondria in cancer therapy. Biochim Biophys Acta 1807(6): 689-696.
- Koppenol, W. H., Bounds, P. L. and Dang, C. V. (2011). Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer 11(5): 325-337.
- Li, Z. and Graham, B. H. (2012). Measurement of mitochondrial oxygen consumption using a Clark electrode. Methods Mol Biol 837: 63-72.
- Plitzko, B., Kaweesa, E. N. and Loesgen, S. (2017). The natural product mensacarcin induces mitochondrial toxicity and apoptosis in melanoma cells. J Biol Chem 292(51): 21102-21116.
- Serill, J. D., Tan, M., Fotso, S., Sikorska, J., Kasanah, N., Hau, A. M., McPhail, K. L., Santosa, D. A., Zabriskie, T. M., Mahmud, T., Viollet, B., Proteau, P. J. and Ishmael, J. E. (2015). Apoptolodins A and C activate AMPK in metabolically sensitive cell types and are mechanistically distinct from oligomycin A. Biochem Pharmacol 93(3): 251-256.
- TeSlaa, T. and Teitell, M. A. (2014). Techniques to monitor glycolysis. Methods Enzymol 542: 91-114.
- Wu, M., Neilson, A., Swift, A. L., Moran, R., Tamagnine, J., Parslow, D., Armistead, S., Lemire, K., Orrell, J., Teich, J., Chomicz, S. and Ferrick, D. A. (2007). Multiparameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. Am J Physiol Cell Physiol 292(1): C125-136.
- Zheng, J. (2012). Energy metabolism of cancer: Glycolysis versus oxidative phosphorylation (Review). Oncol Lett 4(6): 1151-1157.
Data analysis
Results were initially reviewed using the seahorse XF data viewer which automatically saves data as MS Excel (.xls) file. Graphic and statistical analyses were carried out using GraphPad Prism. The significance of observed differences of the basal bioenergetics of cell lines was evaluated by the non-parametric Kruskal-Wallis test followed by Dunn’s multiple comparison post hoc test. In all cases, P < 0.05 was considered to be significant. Experimental values are reported as mean ± standard deviation (Figure 5) or in a box plot (Figure 6).
Figure 5. Mitochondrial stress test. OCR was measured after mensacarcin was injected (black arrow) in different concentrations to SK-Mel-28 cells followed by consecutive injections of oligomycin (1 μM), FCCP (0.5 μM), and antimycin A (0.5 μM)/rotenone (0.5 μM) (n = 3).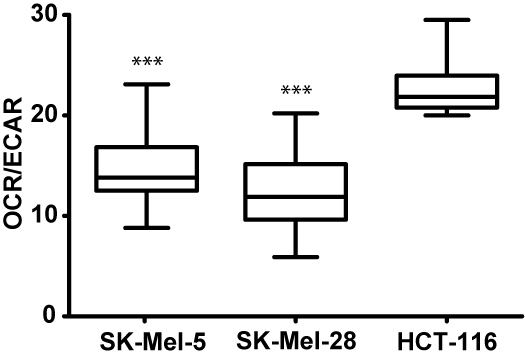
Figure 6. Basal bioenergetic state of SK-Mel-28, SK-Mel-5 and HCT-116 cells. The basal energy metabolism of each cell line was assessed by analyzing OCR/ECAR ratios. OCR and ECAR were acquired with the same protocol as described above but without the injection of compounds. The protocol commands consisted of one loop with 8 measurements. Several separate assays were performed (n = 25).
Notes
Recipes
Acknowledgments
The work was primarily supported by OSU startup funds. B.P. thanks DFG (Deutsche Forschungsgemeinschaft) for postdoctoral funding (PL 799/1-1). We wish to thank Dr. Napur Pande for providing SK-Mel-28 cells, and Bioviotica (Prof. Dr. Axel Zeeck and Hans-Peter Kroll) for providing mensacarcin. We like to thank Elizabeth N. Kaweesa for help with this protocol and Dr. Jeffrey D. Serrill and Prof. Jane E. Ishmael for information and feedback on the seahorse experiments. The authors declare that there are no conflicts of interest or competing interests.
References
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Plitzko, B. and Loesgen, S. (2018). Measurement of Oxygen Consumption Rate (OCR) and Extracellular Acidification Rate (ECAR) in Culture Cells for Assessment of the Energy Metabolism. Bio-protocol 8(10): e2850. DOI: 10.21769/BioProtoc.2850.
- Plitzko, B., Kaweesa, E. N. and Loesgen, S. (2017). The natural product mensacarcin induces mitochondrial toxicity and apoptosis in melanoma cells. J Biol Chem 292(51): 21102-21116.
Category
Cancer Biology > Cellular energetics > Cell biology assays > Metabolism
Cell Biology > Cell metabolism > Other compound
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link