- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Heterologous Expression and Purification of the CRISPR-Cas12a/Cpf1 Protein
Published: Vol 8, Iss 9, May 5, 2018 DOI: 10.21769/BioProtoc.2842 Views: 21739
Reviewed by: Renate WeizbauerRainer MelzerPeter E Burby

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

H2 Production from Methyl Viologen–Dependent Hydrogenase Activity Monitored by Gas Chromatography
Nuttavut Kosem
Dec 5, 2023 1758 Views

Monitoring Protein Stability In Vivo Using an Intein-Based Biosensor
John S. Smetana [...] Christopher W. Lennon
Apr 20, 2025 1574 Views

Endo-1,4-β-D-xylanase Assay Using Azo-Xylan and Variants Thereof
Luca Bombardi [...] Salvatore Fusco
Apr 20, 2025 1916 Views
Abstract
This protocol provides step by step instructions (Figure 1) for heterologous expression of Francisella novicida Cas12a (previously known as Cpf1) in Escherichia coli. It additionally includes a protocol for high-purity purification and briefly describes how activity assays can be performed. These protocols can also be used for purification of other Cas12a homologs and the purified proteins can be used for subsequent genome editing experiments. 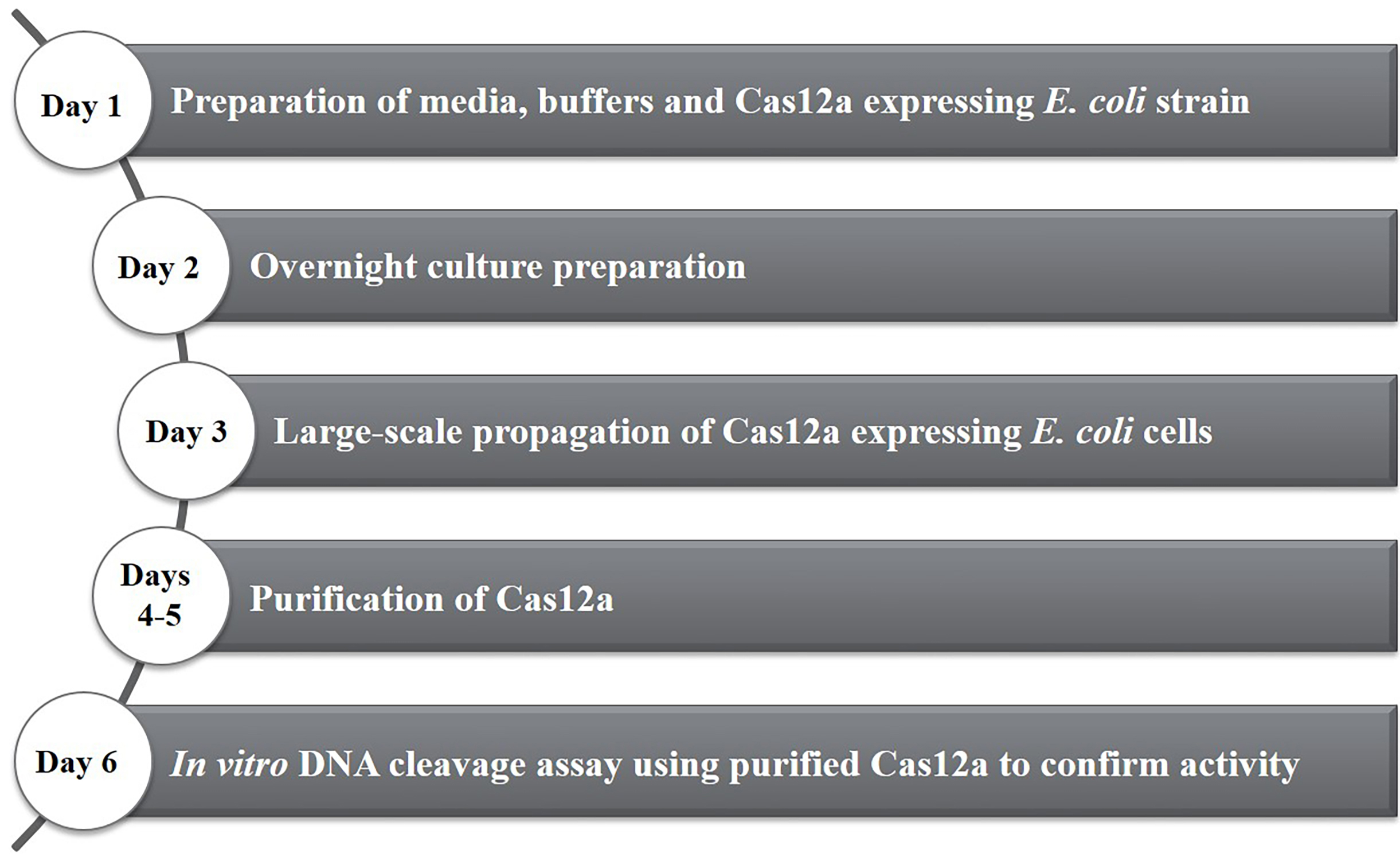
Figure 1. Timeline of activities for the heterologous expression and purification of Francisella novicida Cas12a (FnCas12a) from Escherichia coli
Background
Prokaryotic CRISPR-Cas immune systems provide protection against viruses and plasmids by using CRISPR RNAs (crRNAs) as a guide for sequence-specific targeting of foreign DNA or RNA (van der Oost et al., 2014; Marraffini, 2015). Class 1 CRISPR-Cas systems (comprising types I, III, and IV) typically form multi-subunit protein-crRNA effector complexes, while the class 2 systems (comprising types II, V, and VI) rely on single crRNA-guided effector nucleases for target interference (Mohanraju et al., 2016).
Effector nuclease enzymes from the Class 2 CRISPR-Cas systems have emerged as efficient and precise tools for genome editing and gene expression control (Mali et al., 2013; Doudna and Charpentier, 2014; Hsu et al., 2014). The widely used Cas9, which is the signature protein of type II systems, utilizes a dual guide RNA structure consisting of crRNA and a trans-activating crRNA (tracrRNA) for target recognition (Deltcheva et al., 2011). For genome editing purposes, the dual guide RNA is often replaced by a synthetic fusion of the mature crRNA and tracrRNA, resulting in a long single-molecule guide RNA (sgRNA) in which the individual RNAs are fused by a short linker sequence (Jinek et al., 2012). The sequence of the guide RNA allows binding of complementary DNA targets by base pairing with the target strand, while the other strand of the DNA is displaced. Upon finding a cognate DNA target, the HNH and RuvC nuclease domains of Cas9 mediate cleavage of the target and the displaced strand, respectively (Jinek et al., 2012; Karvelis et al., 2013).
More recently, another novel class 2 CRISPR-Cas nuclease with distinctive features has been identified in bacterial genomes: Cas12a (also known as Cpf1) (Makarova and Koonin, 2015; Zetsche et al., 2015; Shmakov et al., 2017). Cas12a utilizes a single crRNA guide for DNA targeting; it does not require a tracrRNA, resulting in a shorter gRNA sequence compared to the chimeric single-molecule guide RNAs (sgRNA) used by Cas9. While Cas9 requires RNase III-mediated processing of pre-crRNA or individual expression of sgRNAs for the formation of mature guide RNAs, Cas12a can process its own pre-crRNA. This pre-crRNA processing activity allows for simple multiplexing in Cas12a-mediated genome editing (Wang et al., 2017; Zetsche et al., 2017). Whereas Cas9 generates double stranded DNA breaks (DSBs) that are blunt ended, Cas12a generates staggered-end DSBs (Zetsche et al., 2015). Such overhangs can be utilized for overhang-based cloning (Li et al., 2016; Lei et al., 2017). Moreover, Cas9 typically recognizes a G-rich PAM sequence, while all Cas12a orthologues characterized to date recognize a T-rich PAM sequence (Zetsche et al., 2015). Taken together, these features make Cas12a a valuable addition to the genome editing toolbox.
Cas12a has been successfully repurposed for genome editing applications in mammalian cells (Zetsche et al., 2015; Kim et al., 2016a), mice (Hur et al., 2016; Kim et al., 2016b), rice (Endo et al., 2016; Hu et al., 2017; Xu et al., 2017), yeast (Verwaal et al., 2017; Swiat et al., 2017), zebrafish, xenopus (Moreno-Mateos et al., 2017), microalga (Ferenczi et al., 2017) and plant cells (Zaidi et al., 2017; Kim et al., 2017; Tang et al., 2017). The high efficiency and specificity of Cas12a in human cells, coupled with fewer off-target cleavage events compared to Cas9 (Kleinstiver et al., 2016), makes Cas12a a robust and reliable tool for genome editing.
For its in vitro characterization and crystallization (Swarts et al., 2017), Cas12a from Francisella novicida U112 was purified after heterologous expression in Escherichia coli. The expression strain E. coli RosettaTM 2 (DE3) carries a chromosomal T7 RNA polymerase gene under control of an IPTG inducible lacUV5 promoter. The cas12a gene is expressed using a pET vector (Studier and Moffatt, 1986; Rosenberg et al., 1987; Studier et al., 1990) with a lacI-controlled T7 promoter. Here we describe the steps required for controlled expression and purification of FnCas12a. The protocol can also be used for the expression and purification of Cas12a homologs from Acidaminococcus sp. and Lachnospiraceae bacterium.
Materials and Reagents
Note: Equivalent materials and reagents may be used as substitutes.
- Expression of FnCas12a in E. coli RosettaTM 2(DE3)
- 100-ml Erlenmeyer flask (DWK Life Sciences, DURAN®, catalog number: 21 213 24 )
- 2-L Erlenmeyer flask (DWK Life Sciences, DURAN®, catalog number: 21 216 63 )
- 5-L Erlenmeyer flasks (DWK Life Sciences, DURAN®, catalog number: 21 216 73 )
- 50-ml conical centrifuge tubes (Sigma-Aldrich, catalog number: T2318-500EA )
- 2-ml screw top tube (Corning, catalog number: 430659 )
- NalgeneTM PPCO Centrifuge Bottles with Sealing Closure (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 3141-0500 ) or equivalent 500-ml centrifuge bottles
- Pipette tips (DeckWorksTM standard pipet tips, Corning, catalog numbers: 4110 ; 4112 ; 4867 )
- 10-ml syringe (BD, catalog number: 309604 )
- 0.22 μm syringe filter (Mdi, catalog number: SYPL0601MNXX204 )
- 250-ml bottle (Greiner Bio One International, catalog number: 227261 )
- Escherichia coli RosettaTM 2(DE3) cells (Merck, Novagen, catalog number: 71400 ) [encodes a T7 RNA polymerase gene under control of a lacUV5 promoter]
- Plasmid pDS015* [pET His6 TEV LIC cloning vector (Addgene, catalog number: 29653 ), with F. novicida U112 cas12a gene insert fused to an N-terminal His-tag; expression under the control of a lacI-controlled T7 promoter]
*Note: Acidaminococcus sp. BV3L6 Cas12a (AsCas12a) and Lachnospiraceae bacterium ND2006 Cas12a (LbCas12a) proteins can also be purified using this protocol with expression vectors 6His-MBP-TEV-huAsCpf1 (Addgene, catalog number: 90095 ) and 6His-MBP-TEV-huLbCpf1 (Addgene, catalog number: 90096 ) - Tryptone (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: LP0042B )
- Yeast extract (BD, BactoTM, catalog number: 212720 )
- Sodium chloride (NaCl) (Fisher Scientific, catalog number: S271-10 )
- Sodium hydroxide (NaOH) (Merck, EMD Millipore, catalog number: 106462 )
- Ethanol (Fisher Scientific, catalog number: BP2818500 )
- Chloramphenicol (Fisher Scientific, catalog number: BP904100 )
- Kanamycin sulfate (Thermo Fisher Scientific, catalog number: 11815024 )
- Glycerol (Fisher Scientific, catalog number: BP229-4 )
- IPTG (Fisher Scientific, catalog number: BP1755-1 )
- Agar (Acros Organics, catalog number: 400400050 )
- LB medium (see Recipes)
- 1,000x chloramphenicol solution (34 mg/ml) (see Recipes)
- 1,000x kanamycin solution (50 mg/ml) (see Recipes)
- 1 M IPTG (IsoPropyl-1-Thio-β-D-Galactopyranoside) (see Recipes)
- Glycerol stock (50% solution) (see Recipes)
- 100-ml Erlenmeyer flask (DWK Life Sciences, DURAN®, catalog number: 21 213 24 )
- Purification of FnCas12a
- 5 ml HisTrap HP (GE Healthcare, catalog number: 17524701 )
- Dialysis tubing, high retention seamless cellulose tubing, avg. flat width 23 mm (0.9 in.), MWCO 12,400, 99.99% retention (Sigma-Aldrich, catalog number: D0405 )
- Dialysis tubing clamps (Sigma-Aldrich, catalog number: Z371092 )
- 5 ml HiTrap Heparin HP (GE Healthcare, catalog number: 17040601 )
- Amicon Ultra-15 Centrifugal Filter Unit with Ultracel-100 membrane (Merck, EMD Millipore, catalog number: UFC9100 )
- HiLoad 16/600 Superdex 200 pg (GE Healthcare, catalog number: 28989335 )
- GosselinTM Round-Base 10-ml Test Tubes (Corning, GosselinTM, catalog number: TP10-01 ) or other equivalent fraction collection tubes
- NalgeneTM Oak Ridge High-Speed Centrifuge Tubes (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 3114-0050 ) or equivalent 50-ml centrifuge tubes
- Membrane Filter, mixed cellulose esters (Merck, MF-Millipore, catalog number: HAWP04700 )
- Membrane Filter, mixed cellulose esters (Merck, MF-Millipore, catalog number: GSWP04700 )
- Cell pellet from overnight culture in which FnCas12a was expressed (from Procedure A)
- cOmpleteTM, EDTA-free Protease Inhibitor Cocktail (Sigma-Aldrich, Roche Diagnostics, catalog number: 11873580001 )
- Lysozyme from chicken egg white (Sigma-Aldrich, catalog number: L6876-5G )
- β-Mercaptoethanol (Sigma-Aldrich, catalog number: M6250 )
- TEV protease (Sigma-Aldrich, catalog number: T4455 )
- 12% Mini-PROTEAN® TGXTM Precast Protein Gels (Bio-Rad Laboratories, catalog number: 4561043 )
- 4x Laemmli protein sample buffer for SDS-PAGE (Bio-Rad Laboratories, catalog number: 1610747 )
- Bio-SafeTM Coomassie Stain (Bio-Rad Laboratories, catalog number: 1610786 )
- PageRuler Prestained Protein Ladder, 10 to 250 kDa (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 26619 )
- Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: D0632 )
- Ethylenedinitrilotetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: E9884 )
- Sodium chloride (NaCl) (Fisher Scientific, catalog number: S271-10 )
- Tris (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 17926 )
- Imidazole (Sigma-Aldrich, catalog number: I0250 )
- Hydrochloric acid (HCl) (Sigma-Aldrich, catalog number: 258148 )
- Potassium chloride (KCl) (Merck, EMD Millipore, catalog number: 104933 )
- HEPES (Sigma-Aldrich, catalog number: H3375 )
- Potassium hydroxide (KOH) (Sigma-Aldrich, catalog number: 757551 )
- Glycine (Sigma-Aldrich, catalog number: G8898 )
- Sodium dodecyl sulfate (SDS) (Sigma-Aldrich, catalog number: L3771 )
- 1 M DTT (Dithiothreitol) stock (see Recipes)
- 0.5 M EDTA (Disodium Ethylene Diamine Tetra-Acetate) stock (pH 8) (see Recipes)
- Lysis Buffer (see Recipes)
- Wash Buffer (see Recipes)
- Elution Buffer (see Recipes)
- Dialysis Buffer (see Recipes)
- Dilution Buffer (see Recipes)
- IEX-A Buffer (see Recipes)
- IEX-B Buffer (see Recipes)
- SEC Buffer (see Recipes)
- 10x SDS-PAGE Electrophoresis Running Buffer (see Recipes)
- 5 ml HisTrap HP (GE Healthcare, catalog number: 17524701 )
- Activity assay using purified Cas12a
- Purified Cas12a Nuclease (from Procedure B)
- Nuclease-free water
- Proteinase K, Molecular Biology Grade (New England Biolabs, catalog number: P8107S )
- crRNA containing the targeting sequence complementary to the target DNA
Note: The RNA can be ordered as a desalted RNA oligonucleotide or as PAGE-purified RNA oligonucleotide from an RNA synthesis company such as Sigma-Aldrich or IDT. - DNA substrate containing the target sequence and a 5’ TTTN PAM sequence
Note: The substrate DNA can be circular or linearized plasmid, PCR products, or synthesized oligonucleotides). As an example, the DNA substrate and crRNA used in the activity assay is shown in Figure 2.
Figure 2. Schematic of the Cas12a crRNA-DNA-targeting complex. The expected cleavage sites are indicated by red arrows. - GeneRuler 1 kb DNA Ladder (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: SM0311 ) or equivalent
- DNA gel Loading Dye [e.g., 6x DNA Loading Dye (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: R0611 )]
- InvitrogenTM SYBRTM Safe DNA Gel Stain (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: S33102 )
- Sodium chloride (NaCl) (Fisher Scientific, catalog number: S271-10 )
- Magnesium chloride hexahydrate (MgCl2·6H2O)
- HEPES (Sigma-Aldrich, catalog number: H3375 )
- Ethylenedinitrilotetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: E9884 )
- Hydrochloric acid (HCl) (Sigma-Aldrich, catalog number: 258148 )
- 10x Nuclease Reaction Buffer (see Recipes)
- Purified Cas12a Nuclease (from Procedure B)
Equipment
Note: Equivalent equipment can be used.
- Expression of FnCas12a in E. coli RosettaTM 2
- Pipettes (Corning, model: LambdaTM Plus Single-Channel Pipettor, catalog numbers: 4070 ; 4074 ; 4075 )
- New BrunswickTM Innova® 42 incubator (Eppendorf, New BrunswickTM, model: Innova® 42 , catalog number: M1335-0002) or an equivalent incubator that can be set at 37 °C
- Sorvall LYNX 4000 Superspeed Centrifuge (Thermo Fisher Scientific, Thermo ScientificTM, model: Sorvall LYNX 4000 , catalog number: 75006580) or an equivalent centrifuge that can be cooled down to 4 °C and can perform up to 6,000 x g
- New BrunswickTM Innova® 44/44R (Eppendorf, New BrunswickTM, model: Innova® 44/44R , catalog number: M1282-0002) or any equivalent shaker incubator where the temperature can be set at 37 °C and 18 °C
- Cell density meter (GE Healthcare, model: UltrospecTM 10 , catalog number: 80-2116-30), or equivalent spectrophotometer that can measure the density of cells in suspension at 600 nm
- Ice-water bath (water and ice mixed)
- Pipettes (Corning, model: LambdaTM Plus Single-Channel Pipettor, catalog numbers: 4070 ; 4074 ; 4075 )
- Purification of FnCas12a
- SONOPULS HD (Bandelin electronic, model: HD 3200 ) with VS 70 T Sonotrode (Bandelin) or equivalent ultrasonic homogenizer/Sonifier, or alternatively a French Pressure Cell (French Press) for cell lysis
- Peristaltic pump P-1 with connectors for 5 ml HisTrap HP (GE Healthcare, model: Peristaltic Pump P-1, catalog number: 18111091 ) or an equivalent peristaltic pump
- Tubing Connectors for Use with Peristaltic Pump P-1 (GE Healthcare, catalog number: 11300082 )
- ÄKTApurifier 10 FPLC system (GE Healthcare, model: ÄKTApurifier 10 , catalog number: 28406264) or an equivalent FPLC system
- Sorvall LYNX 4000 Superspeed Centrifuge (Thermo Fisher Scientific, Thermo ScientificTM, model: Sorvall LYNX 4000 , catalog number: 75006580) or an equivalent centrifuge that can be cooled down to 4 °C and can perform up to 30,000 x g
- pH meter (QiS, model: B210 )
- Filter holder assembly for filtration (Merck, catalog number: XX1014700 or Nalgene, Thermo Fisher Scientific, Thermo ScientificTM, catalog number: DS0320-2545 ), or equivalent filter holder assembly
- Diaphragm Vacuum Pumps LABOPORT® N 820 (ABM van Zijl B.V, catalog number: ABMK N8203FT18 ), or an equivalent vacuum pump
- Nanodrop (Thermo Fisher Scientific, Thermo ScientificTM, model: NanoDropTM 2000 , catalog number: ND-2000)
- Mini-PROTEAN Tetra cell (Bio-Rad Laboratories, model: Mini-PROTEAN Tetra Cell, catalog number: 1658004EDU ), or an equivalent vertical electrophoresis system
- Epson Perfection V850 Pro scanner (Epson, model: Perfection V850 Pro ) or equivalent scanner or imager suitable for SDS-PAGE gel imaging.
- SONOPULS HD (Bandelin electronic, model: HD 3200 ) with VS 70 T Sonotrode (Bandelin) or equivalent ultrasonic homogenizer/Sonifier, or alternatively a French Pressure Cell (French Press) for cell lysis
- Activity assay using purified Cas12a
- EppendorfTM 5424 Microcentrifuge (Eppendorf, model: 5424 , catalog number: 022620498)
- MUPID One Horizontal Electrophoresis System (Bulldog Bio, catalog number: MU2 ) or an equivalent horizontal electrophoresis system
- G:BOX F3 (Syngene, model: G:BOX F3 , catalog number: 05-GBOX-F3) gel doc system or equivalent DNA agarose gel imaging equipment
- EppendorfTM 5424 Microcentrifuge (Eppendorf, model: 5424 , catalog number: 022620498)
Procedure
- Transformation of E. coli RosettaTM 2(DE3) with pDS015 plasmid and preparation of a glycerol stock
- Add 1 ng of pDS015 plasmid DNA directly to 50 µl of chemically competent E. coli RosettaTM 2(DE3) cells.
- Stir gently to mix and place the tubes on ice for 5 min.
- Heat the tubes for exactly 30 sec in a 42 °C water bath; do not shake.
- Immediately place the tube on ice for 2 min.
- Add 250 μl of room temperature sterile SOC medium (provided by the manufacturer) to the tube. Incubate at 37 °C while shaking at 250 rpm for 60 min.
- Spread 5-50 μl of the transformation mixture on LB agar plates containing 34 µg/ml and 50 µg/ml of chloramphenicol and kanamycin, respectively. If plating less than 25 μl of the transformation, we recommend adding 50 µl of sterile SOC medium to the transformation mixture before plating to facilitate even colony distribution on the LB agar plate surface.
- Incubate the LB agar plates overnight at 37 °C.
- The next day, pick a single colony from the transformation plates using a sterile pipette tip and inoculate 10 ml LB in a 50-ml tube.
- Incubate the 50-ml tube overnight in a 37 °C shaking incubator shaking at 160 rpm.
- The next day, add 500 μl of the overnight culture to 500 μl of 50% sterile glycerol in a 2-ml screw-top tube or cryovial and mix gently.
- Store the glycerol stock at -80 °C for future use.
- Add 1 ng of pDS015 plasmid DNA directly to 50 µl of chemically competent E. coli RosettaTM 2(DE3) cells.
- Large-scale expression of FnCas12a in E. coli RosettaTM 2(DE3)
Day 1. Preparation of media, buffers and single colonies- Prepare 20 ml of LB in a 100-ml Erlenmeyer flask (for starting overnight cultures) (Recipe A1).
- Prepare three 5-L Erlenmeyer flasks, each containing 1.5 L of LB medium (for large-scale propagation and protein purification) (Recipe A1).
- Prepare antibiotic and stock solutions (Recipes A2 and A3).
- Prepare the buffers needed for the purification (Recipes B1-B8).
- Streak out a glycerol stock of E. coli RosettaTM 2(DE3) transformed with pDS015 on an LB agar plate containing 50 μg/ml kanamycin and 34 μg/ml chloramphenicol.
- Incubate the LB agar plate overnight in a 37 °C incubator.
Day 2. Overnight culture preparation- Add 20 μl of the 50 mg/ml kanamycin stock solution and 20 μl of the 34 mg/ml chloramphenicol stock solution to the 20 ml of autoclaved LB medium in a 100-ml Erlenmeyer flask from Day 1.
- With a sterile pipette tip, pick a single colony of E. coli RosettaTM 2(DE3) transformed with pDS015 from the LB agar plate from Day 1.
- Use the colony to inoculate the medium containing the kanamycin and chloramphenicol.
- Loosely close the 100-ml Erlenmeyer flask with a cotton plug.
- Incubate the bacterial culture at 37 °C for 16-20 h in a shaking incubator (set at 160 rpm).
Day 3. Large-scale propagation of cells overexpressing FnCas12a- Take three autoclaved 5-L Erlenmeyer flasks* each containing 1.5 L LB medium from Day 1.
- To each flask, add 1.5 ml of 50 mg/ml chloramphenicol solution.
- To each flask, add 1.5 ml of 34 mg/ml kanamycin solution.
- To each flask, add 15 ml of the overnight culture prepared on Day 2.
- Incubate the culture flasks at 37 °C in a shaking incubator at 160 rpm**.
- Monitor the OD600 nm of the culture every half an hour. Once an OD600 nm of 0.5-0.6 is reached (this normally takes ~3-4 h), transfer the Erlenmeyer containing the culture to the ice-water bath and incubate (cold-shock) it for 15 min. This step slows down the metabolism of E. coli and triggers expression of cold-shock proteins which may aid FnCas12a folding during expression.
- To each flask, add 200 μl of filter-sterilized 1 M IPTG solution to the culture to induce expression of FnCas12a.
- Transfer the culture to an 18 °C shaking incubator (set at 120 rpm) for overnight expression (~16 h).
Notes:- *It is also possible to express smaller volumes of cell culture (e.g., a single 1.5 L culture or one or more 750 ml cultures in 2-L Erlenmeyer flasks)
- **When using a baffled Erlenmeyer flask, reduce the shaking incubator speed to 120 rpm to prevent the formation of foam.
- *It is also possible to express smaller volumes of cell culture (e.g., a single 1.5 L culture or one or more 750 ml cultures in 2-L Erlenmeyer flasks)
Day 4–Part I. Large-scale propagation of cells overexpressing FnCas12a (continued)- Transfer the overnight culture from Day 3 to centrifuge bottles.
- Harvest the cells by centrifuging the culture for 15 min at 6,000 x g at 4 °C.
- Discard supernatant and store the pelleted cells at -20 °C (for use within a week for optimal purification) or at -80 °C (for long-term storage) or proceed directly to purification. The expected yield is ~5 g of cell pellet per liter of cell culture.
Day 4–Part II. Purification of FnCas12a–Part I- If continuing with a frozen cell pellet, thaw the cell pellet from Day 4–Part I on ice for 30-60 min. If proceeding directly after protein expression, skip this step.
Note: All subsequent steps should be performed on ice or at 4 °C. - Resuspend the entire cell pellet in Lysis Buffer (~2.5-5 ml Lysis Buffer per gram of cell pellet).
- Add 1 tablet cOmpleteTM protease inhibitor for every 50 ml.
- Add lysozyme to a final concentration of 1 mg/ml.
- Incubate the sample on ice for 30 min.
- If using a French Press for cell lysis: after the lysozyme treatment, pass cell suspension through French Press twice at 16,000 psi.
- If using sonication for cell lysis: after the lysozyme treatment, lyse the cell suspension by using a sonicator with an appropriate tip and a protocol suitable for lysis of large volume cell suspensions. For our setup (Bandelin SONOPULS HD with VS 70 T tip), we use the following settings: 10 min total time, 1 sec on, 0.7 sec off, and 20% amplitude. The cell suspension often has a brownish tinge after lysis as shown in Figure 3.
Note: Using a French Pressure cell or sonicator gives approximately the same yield–FnCas12a is very stable and little to no protein will be lost during sonication. Keep in mind, however, that sonication is usually less suitable for large volumes, and therefore a protocol suitable for lysis of a large volume of cell suspension should be applied.
Figure 3. After lysis, the cell suspension becomes tinted brown and less viscous - Pour the lysate into (a) centrifugation tube(s) and centrifuge for 45 min, 4 °C, at 30,000 x g.
- Transfer the supernatant to (a) clean 50-ml tube(s). This is the ‘cell-free extract’.
Note: A sample of the lysed cell pellet may be stored at 4 °C and analyzed later by SDS-PAGE analysis for the presence of FnCas12a to assess its solubility and to determine if cell lysis was successful. - Pass the cell free extract through a 0.22 µm membrane filter and save the filtrate into a sterile tube.
- Using a peristaltic pump, wash a HisTrap HP column with 3-5 column volumes of distilled water to remove the solution in which the resin is stored.
- Using a peristaltic pump, equilibrate the HisTrap HP with at least 5 column volumes of Lysis Buffer with a flow rate of 2 ml/min.
- Using the peristaltic pump*, pass the filtered cell free extract with a flow rate of 1 ml/min through the HisTrap HP column and collect the flow-through in (a) 50-ml tube(s) labeled ‘flow-through’**.
Notes:- *For sample loading, a superloop can be used instead of the peristaltic pump.
- **A sample of the flow-through may be stored at 4 °C and analyzed later by SDS-PAGE analysis for the presence of the FnCas12a to determine if the protein bound to the column (if a large fraction of the protein remains in the flow-through, regenerate or replace your column). In some cases, the high amount of proteins can saturate the column. To recover the protein in the flow-through, the flow-through can be (re)loaded onto another or regenerated HisTrap HP column.
- *For sample loading, a superloop can be used instead of the peristaltic pump.
- Equilibrate the ÄKTA FPLC system with Wash Buffer until the absorbance at 280 nm reaches a steady baseline. Transfer the HisTrap HP column to the ÄKTA FPLC and wash the column using Wash Buffer with a flow rate of 2 ml/min for 10-15 times the column volume or until the absorbance at 280 nm becomes near** stable with the Wash Buffer. Collect the first 3-4 10-ml wash fractions* in separate tubes labeled ‘wash-through #’.
Notes:- *The wash-through may be saved and checked later using SDS-PAGE analysis for the presence of FnCas12a to determine if it was eluted off the column during washing.
- **Even at its low concentration, imidazole in the washing buffer can remove small amounts of the protein of interest. Therefore, start eluting the protein as the absorbance at 280 nm is near stable to avoid unnecessary loss of the protein of interest.
- *The wash-through may be saved and checked later using SDS-PAGE analysis for the presence of FnCas12a to determine if it was eluted off the column during washing.
- Elute the protein using the Elution Buffer with a flow rate of 2 ml/min while fractionating to 1 ml samples, collect the eluate and save the fractions in separate tubes labeled ‘Elution fraction #’. An example of a typical elution chromatogram of FnCas12a purified by Histrap HP (5 ml) affinity purification is shown in Figure 4.
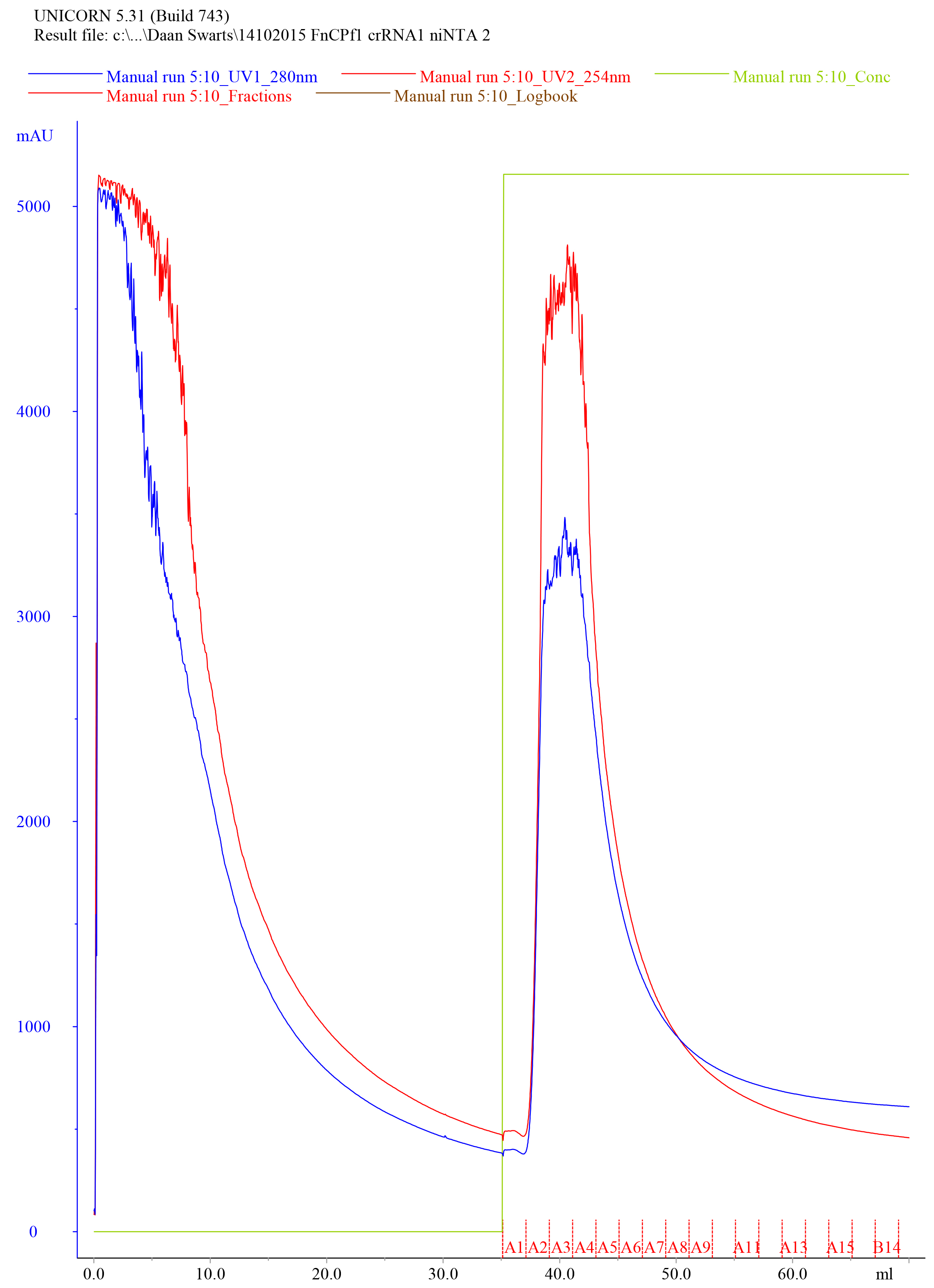
Figure 4. Representative elution chromatogram of FnCas12a purified by Histrap HP (5 ml) affinity purification. 75 ml of cell-free extract was loaded on the column. Elution fractions were 1 ml and the flow rate was set at 2 ml/min. Absorbance at 280 nm is expressed in milli-absorbance units for the A280 (blue) and A254 (red). Please note that the A254 is not very informative after niNTA purification, as at this stage, the sample is contaminated with various nucleic acids. The green line indicates the concentration of Elution Buffer (0% at the start of the chromatogram and 100% at the final stage of the chromatogram). - Dilute 10 µl of the collected fractions with 4x Laemmli protein sample buffer, heat for 5 min at 95 °C and resolve the samples on SDS-PAGE gel to assess the purity of the sample.
- Combine the elution fractions in which the protein is present and check the absorbance at 280 nm to estimate the protein concentration. The extinction coefficient of FnCas12a is 145,820 M-1 cm-1. Typical yield at this step is ~25 mg per liter of expression culture**.
Note: **The yield is most likely overestimated at this point due to protein and nucleic acid contaminations. - Add 2 ml (final 1 mM) of 1 M DTT stock and 4 ml of 0.5 M EDTA stock to 2 L of Dialysis Buffer before use. Increase the volume of the combined protein fractions to 25 ml using the Dialysis Buffer. Add 1 mg TEV per 100 mg of protein.
- Take a dialysis membrane with an MWCO of 12,400 and soak it in the Dialysis Buffer for 1 min. Use a clamp to close the dialysis membrane on one end to make a bag.
- Pipet the combined protein sample into the dialysis membrane bag and close the other end with another clamp. Dialyze the sample overnight at 4 °C with slow stirring against 2 L of the Dialysis Buffer.
Day 5. Purification of FnCas12a–Part II- Transfer the protein sample from the dialysis membrane into a 50 ml tube and centrifuge it for 10 min at 4,500 x g at 4 °C to remove potential precipitated proteins.
- Dilute the sample 1:1 using Sample Dilution Buffer*.
- Using a peristaltic pump, wash the column with 3-5 column volumes of distilled water to remove the solution in which the column resin is stored.
- Using a peristaltic pump, equilibrate the Heparin FF column with at least 5 column volumes of IEX-A Buffer with a flow rate of 2 ml/min.
- Using a peristaltic pump, load the protein sample onto the Heparin FF column.
- Equilibrate the ÄKTA FPLC with IEX-A Buffer until the absorbance at 280 nm reaches a steady baseline.
- Transfer the Heparin FF column to the ÄKTA FPLC and wash it with 10 ml IEX-A Buffer at 2 ml/min. Collect the flow-through in appropriately labeled clean tubes.
- Elute the protein using a linear gradient from 0 to 50% IEX-B Buffer over 60 ml at a flow rate of 2 ml/min, collect the eluate as 1 ml fractions in appropriately labeled clean tubes. An example of a typical elution chromatogram of FnCas12a purified by Heparin FF (5 ml) affinity purification is shown in Figure 5.
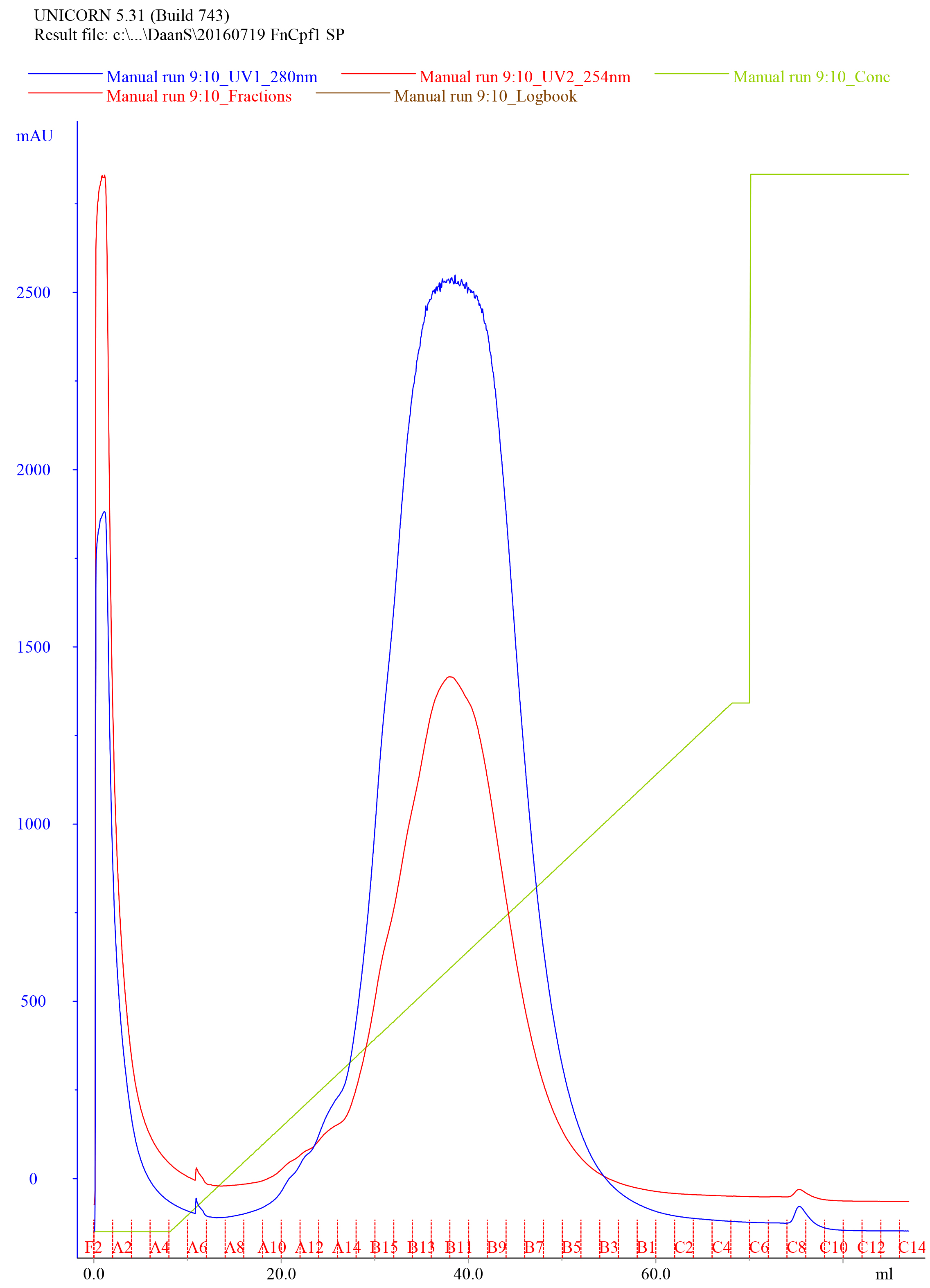
Figure 5. Representative elution chromatogram of FnCas12a purified by Heparin FF (5 ml) affinity purification. 50 ml of sample was loaded. The flow rate was set at 2 ml/min and elution fractions were 1 ml. Absorbance is expressed in milli-absorbance units for the A280 (blue) and A254 (red). The green line indicates the concentration of IEX-B (0% at the start of the chromatogram with the concentration raising to 50% over 60 ml, and at 100% at the final stage of the chromatogram to wash the column). - Dilute 10 µl of the collected fractions with 4x Laemmli protein sample buffer, heat for 5 min at 95 °C, and analyze on SDS-PAGE gel to assess the purity of the sample. An example of a Coomassie Brilliant Blue stained 10% SDS-PAGE gel on which FnCas12a Heparin FF elution fractions were resolved is shown in Figure 6.
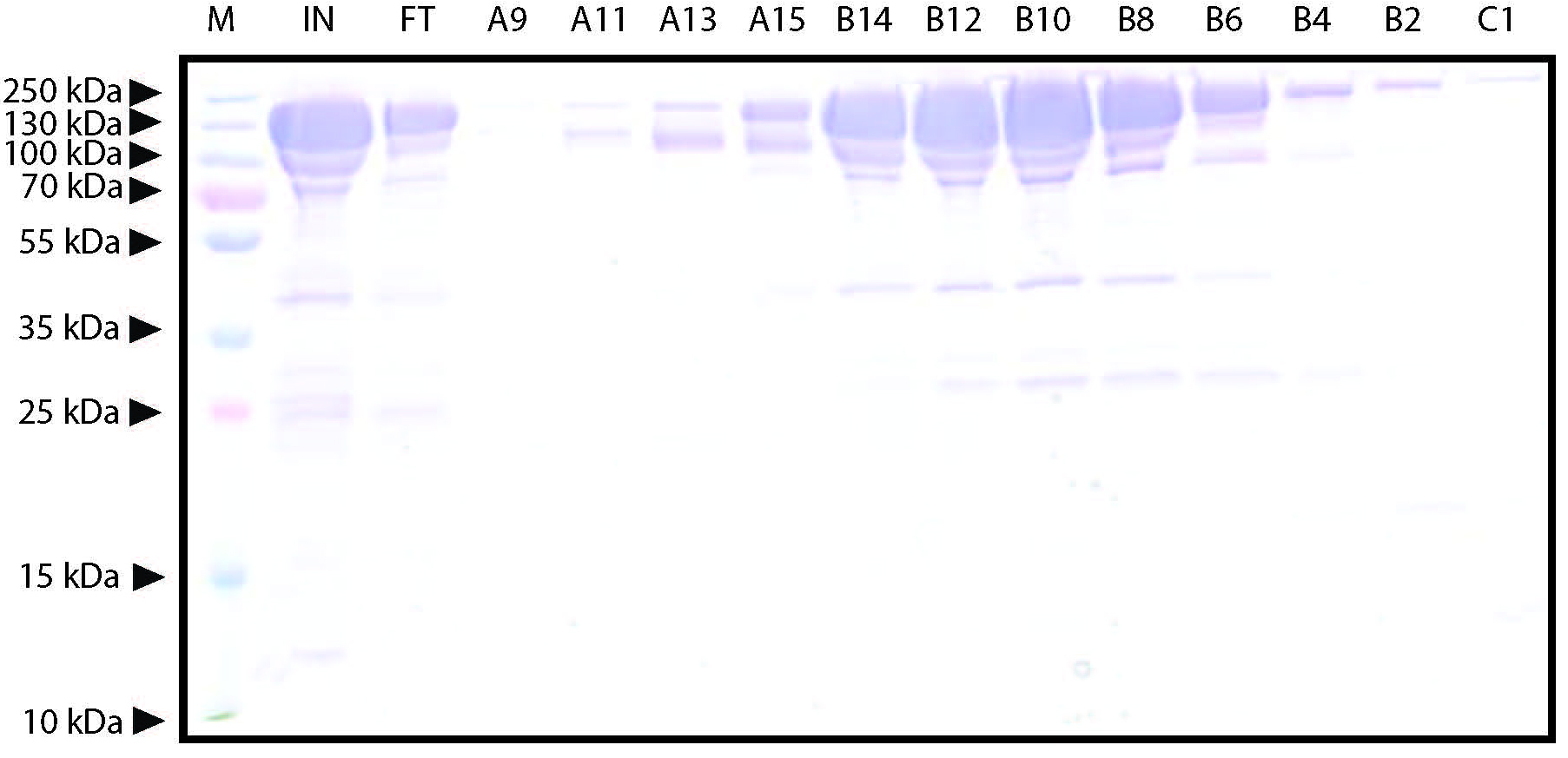
Figure 6. Representative Coomassie Brilliant Blue stained 12% SDS-PAGE gel on which FnCas12a Heparin FF elution fractions were resolved. M: PageRulerTM Plus Prestained Protein Ladder. Marker band sizes are indicated in kDa. IN: concentrated input sample of TEV protease-treated Histrap HP elution fractions after overnight dialysis. FT: Flow through from the column. Besides the large band formed by FnCas12a, other (contamination) bands can be observed. A9-C1: Elution fractions near the protein absorbance peak. The FnCas12a protein appears as a band with a size slightly larger than 130 kDa. In this case, fractions B14-C1 were combined. - Combine the elution fractions containing pure protein (and as little contaminants as possible) and concentrate the sample by transferring the sample to Amicon Ultra-15 Centrifugal Filter Units with a membrane MWCO of 100 kD and centrifuging the samples at 4,500 x g at 4 °C until a final volume of < 1 ml is reached.
- Transfer the sample to an Eppendorf tube and centrifuge the sample for 2 min at maximum speed in a pre-cooled (4 °C) microcentrifuge to remove potential precipitation.
- Equilibrate a 2-ml injection loop and the HiLoad 16/600 Superdex 200 pg column with 10 ml and 240 ml SEC Buffer, respectively, on the ÄKTA FPLC.
- Load the protein concentrate on the HiLoad 16/600 Superdex 200 pg column using the 2-ml injection loop and resolve the sample on the column using SEC Buffer with a flow rate of 1 ml/min.
- Collect 1 ml fractions. An example of a typical elution chromatogram of FnCas12a purified by HiLoad 16/600 Superdex 200 pg column size exclusion purification is shown in Figure 7.
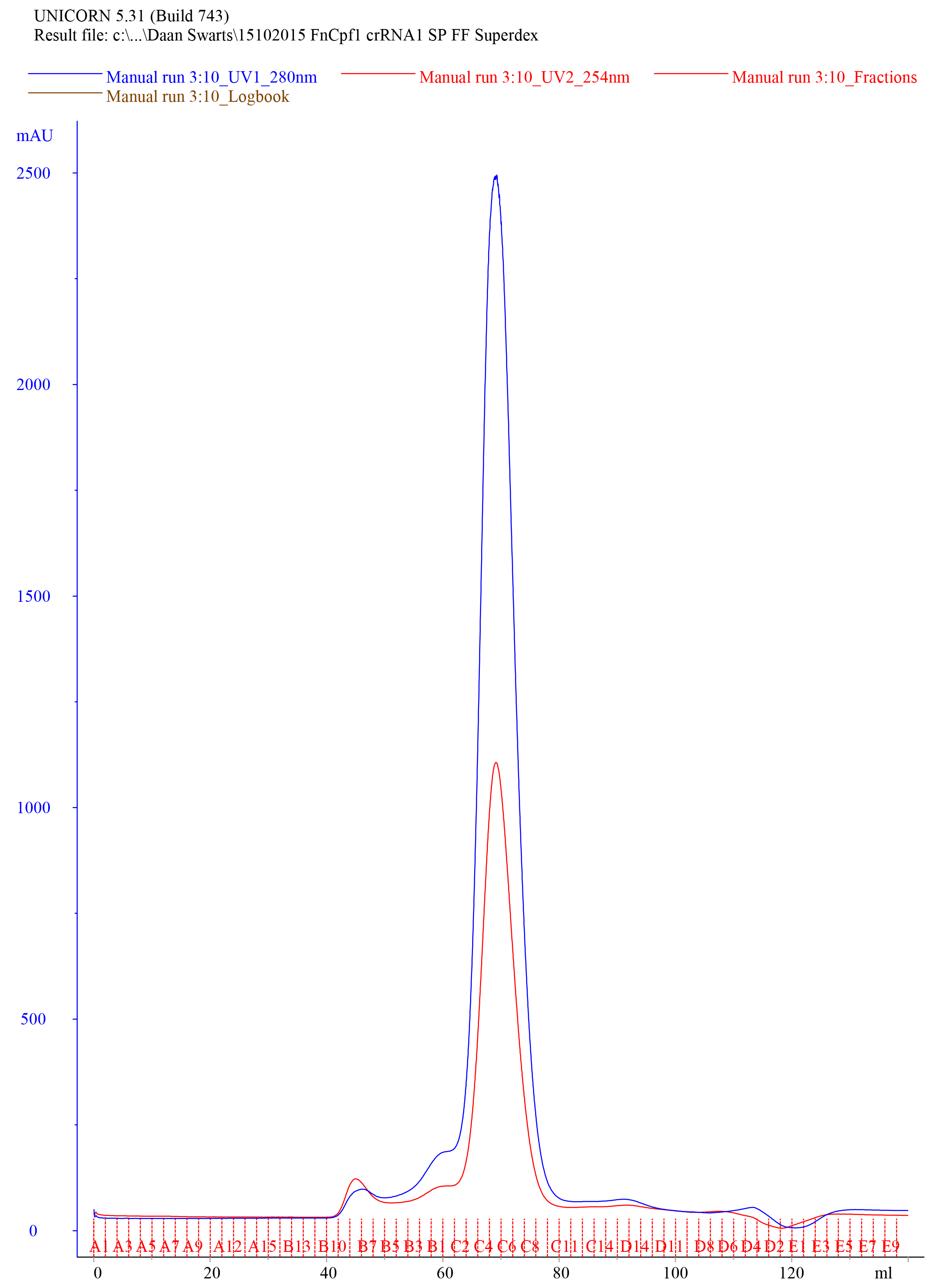
Figure 7. Representative elution chromatogram of FnCas12a resolved on a HiLoad 16/600 Superdex 200 pg column. 1 ml of sample was loaded. Elution fractions were 1 ml and the flow rate was set at 1 ml/min. Absorbance is expressed in milli-absorbance units for the A280 (blue) and A254 (red). - Dilute 10 µl of the collected fractions with 4x Laemmli protein sample buffer, heat for 5 min at 95 °C, and resolve on a 10% SDS-PAGE gel to assess the purity of the sample. An example of a Coomassie Brilliant Blue stained 10% SDS-PAGE gel on which FnCas12a SEC elution fractions were resolved is shown in Figure 8.
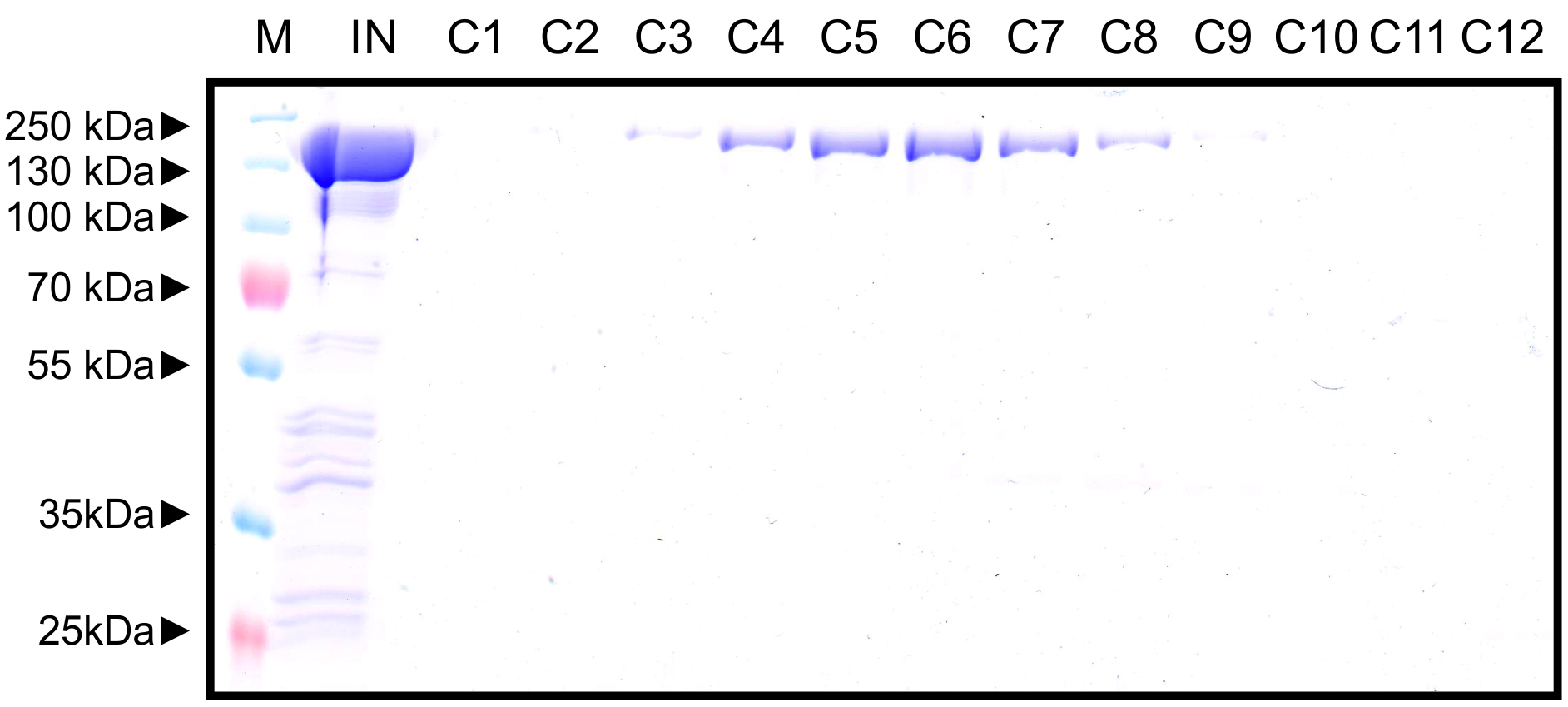
Figure 8. Representative Coomassie Brilliant Blue stained 12% SDS-PAGE gel on which FnCas12a SEC elution fractions were resolved. M: PageRuler Plus Prestained Protein Ladder. Marker band sizes are indicated in kDa. IN: concentrated input sample. Besides the large band formed by FnCas12a, other (contamination) bands can be observed. C1-C12: Elution fractions near the protein absorbance peak. The FnCas12a protein appears as a band with a size slightly larger than 130 kDa. In this case, fractions C3-C9 were combined. - Combine and concentrate the fractions that contain the pure protein and no (or a negligible amount of) contaminants.
- For analysis of purity and final protein yield, see the Data analysis section. The typical protein yield at this step is ~5-10 mg of FnCas12a per liter of E. coli expression culture. Expression and purification of AsCas12a or LbCas12a typically results in slightly higher Cas12a yields.
- Dilute the samples to a concentration suitable for subsequent experiments (e.g., 10 µM)** and aliquot the protein at a desired concentration and store at -80 °C.
Notes:- *For LbCas12a, dilute in a 2:1 ratio (protein sample: Dilution Buffer) due to the instability of LbCas12a at lower salt concentrations.
- **It is recommended to store the protein at ≥ 10 µM and dilute it to the right concentration only just before use. At lower concentrations, a relative high fraction of the protein can be lost due freezing/thawing and non-specific adsorption to the surface of the tube/container used for storage.
- *For LbCas12a, dilute in a 2:1 ratio (protein sample: Dilution Buffer) due to the instability of LbCas12a at lower salt concentrations.
Day 6. In vitro cleavage assay for confirming the activity of purified FnCas12a
Notes:- We strongly recommend wearing gloves and using nuclease-free tubes and reagents to avoid RNase contamination.
- The reaction volume is typically 20 μl but can be scaled up as needed. Reactions should be assembled in nuclease-free 1.5 ml Eppendorf tubes or in 200 µl PCR (strip) tubes.
- Prepare a 1 µM crRNA solution by diluting the stock with nuclease-free water on ice.
- Prepare a 0.1 µM substrate plasmid or linear DNA solution by diluting the stock with nuclease-free water on ice.
- Prepare the following two-step reaction (at a molar ratio Cas12a:crRNA:substrate = 10:20:1) at room temperature:
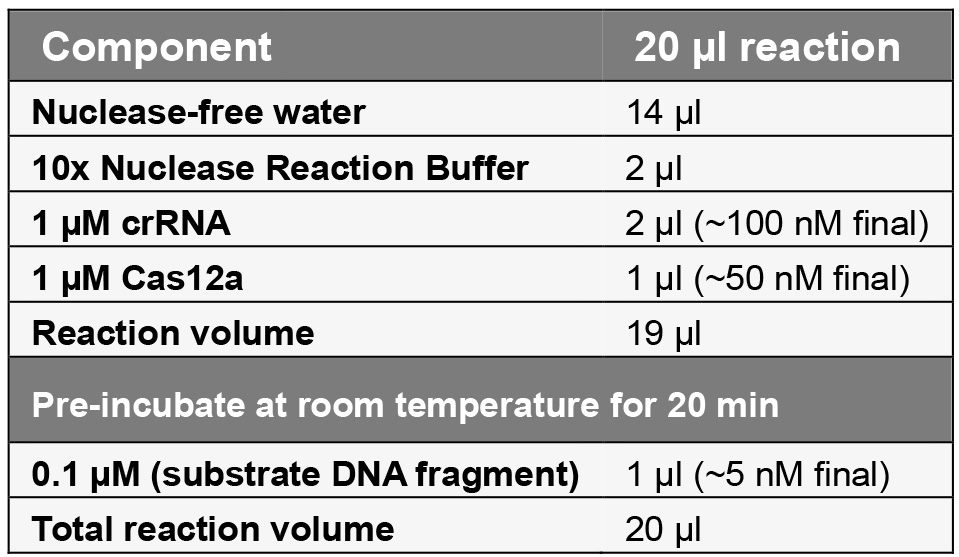
- Mix thoroughly and pulse-spin in a micro-centrifuge.
- Incubate at 37 °C for 30 min.
- Add 1 µl of Proteinase K, mix thoroughly and pulse-spin in a micro-centrifuge.
- Incubate at room temperature for 10 min.
- Add 4 µl of 6x DNA loading dye.
- Resolve 20 µl of the sample on an 1% agarose gel pre-stained with SYBRTM Safe DNA Gel Stain.
- Visualize the gel using an imaging system equipped with an excitation source in the UV range or between 470-530 nm. An example of an in vitro cleavage assay using FnCas12a:crRNA and a linear DNA substrate is shown in Figure 9.
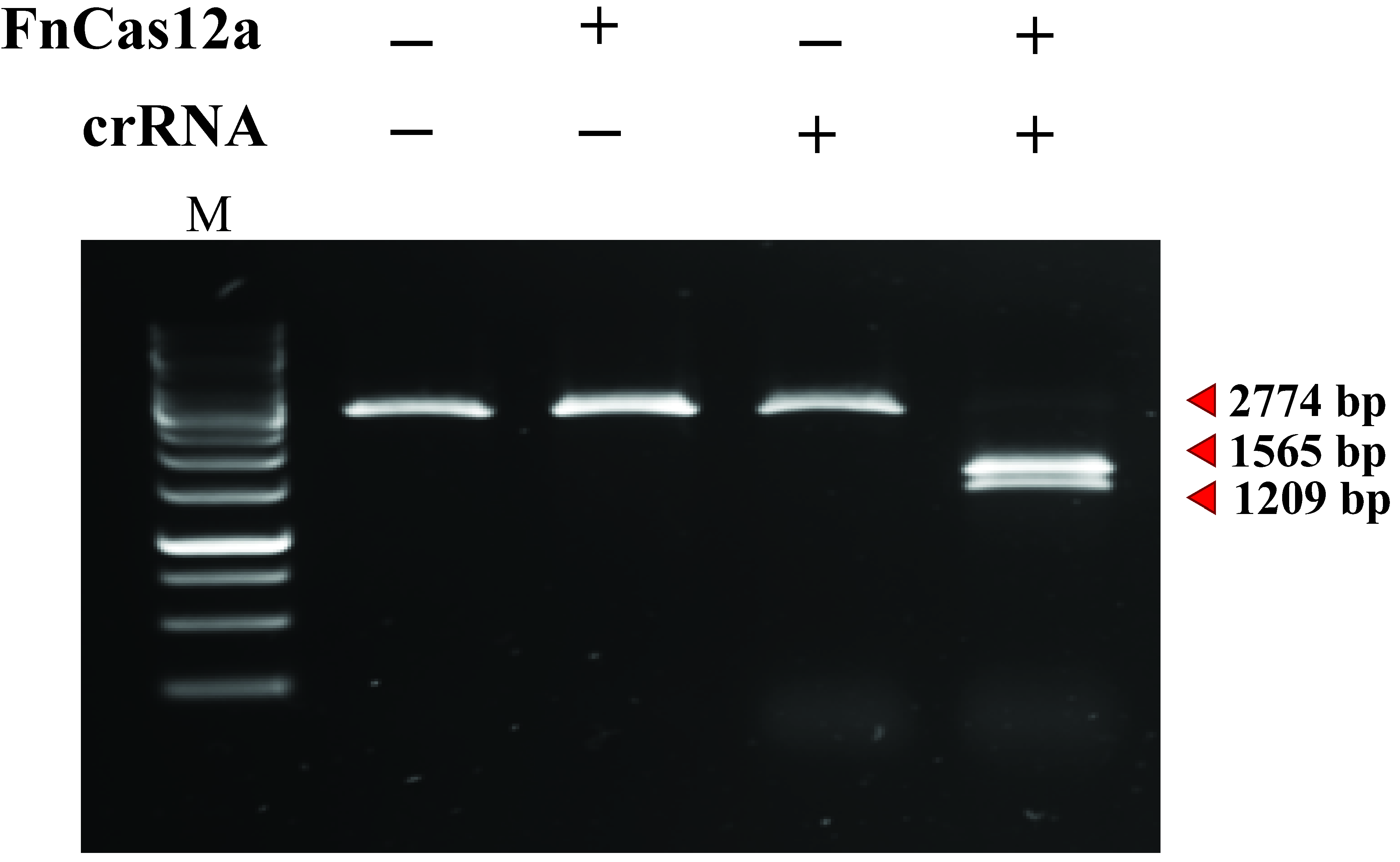
Figure 9. A 2,774 bp linear target DNA substrate is cleaved by the FnCas12:crRNA complex, yielding products of 1,209 bp and 1,565 bp. M: GeneRuler 1 kb DNA Ladder.
- Prepare 20 ml of LB in a 100-ml Erlenmeyer flask (for starting overnight cultures) (Recipe A1).
Data analysis
Note: This section explains how to determine the yield and purity of your protein after the final step of the purification protocol (i.e., after size exclusion chromatography).
- Use a NanoDrop Spectrophotometer to measure the protein concentration. When using the NanoDrop software method ‘Protein A280’, both the absorption at 260 nm and the absorption at 280 nm are measured.
- The 260/280 ratio of the purified protein sample can be used to determine if the complex is free from nucleic acids. Typically, a pure protein sample has a 260/280 ratio of ~0.57. We typically achieve a 260/280 ratio between 0.54 and 0.61. Nucleic acid contamination rapidly increases the 260/280 ratio to above 1.
- Use the measured absorbance at 280 nm to determine the final protein concentration.
- When using the ‘Other protein (ε + MW)’ option of the NanoDrop software, provide the molar extinction coefficient of FnCas12a (144,330 M-1 cm-1) and its molecular weight (151 kDa). For AsCas12a, these values are 15,780 M-1 cm-1 and 156 kDa. For LbCas12a, these values are 181,690 M-1 cm-1 and 149 kDa.
- When using the ‘1 Abs = 1 mg/ml’ option of the NanoDrop software, use a correction factor to determine the real protein concentration. The correction factors for FnCas12a, AsCas12a, and LbCas12a are 0.951, 1.009 and 1.221, respectively. For example, if the NanoDrop measurement gives a protein concentration of 5.0 mg/ml of FnCas12a (assuming 1 Abs = 1 mg/ml), your real protein concentration is 5/0.951 = 5.26 mg/ml. To calculate the protein concentration in mM, use the following formula: protein concentration (in mg/ml)/molecular weight (in kDa; given above). For example, if you have a FnCas12a sample with 5.26 mg/ml protein, the protein concentration is 5.26/151 = 0.035 mM = 35 µM.
- When using the ‘Other protein (ε + MW)’ option of the NanoDrop software, provide the molar extinction coefficient of FnCas12a (144,330 M-1 cm-1) and its molecular weight (151 kDa). For AsCas12a, these values are 15,780 M-1 cm-1 and 156 kDa. For LbCas12a, these values are 181,690 M-1 cm-1 and 149 kDa.
Recipes
- Media, antibiotics and stock solutions
- LB medium (1 L)
- Weigh out 10 g tryptone, 5 g yeast extract, and 10 g NaCl
- Fill up to 800 ml with demi-water
- Adjust pH to 7.5 with NaOH
- Fill up to 1 L with demi-water
- Autoclave at 121 °C for 30 min
- Store at room temperature
- Weigh out 10 g tryptone, 5 g yeast extract, and 10 g NaCl
- 1,000x chloramphenicol stock (34 mg/ml stock)
- Weigh out 0.34 g chloramphenicol and dissolve it in 10 ml of 100% ethanol
- Pass through a 0.22 μm syringe filter
- Store at -20 °C
- Weigh out 0.34 g chloramphenicol and dissolve it in 10 ml of 100% ethanol
- 1,000x kanamycin stock (50 mg/ml stock)
- Weigh out 0.5 g kanamycin sulfate and dissolve it in 10 ml of sterile water
- Pass through a 0.22 μm syringe filter
- Store at -20 °C
- Weigh out 0.5 g kanamycin sulfate and dissolve it in 10 ml of sterile water
- Glycerol stock (50% solution)
- Add 50 ml of 100% glycerol solution into a 250-ml bottle. When pipetting glycerol, use ethanol sterilized scissors to cut off the end of a pipette to make pipetting easier
- Add 50 ml demi-water
- Autoclave at 121 °C for 30 min
- Store at room temperature
- Add 50 ml of 100% glycerol solution into a 250-ml bottle. When pipetting glycerol, use ethanol sterilized scissors to cut off the end of a pipette to make pipetting easier
- 1 M IPTG (IsoPropyl-1-Thio-β-D-Galactopyranoside)
- Weigh out 2.38 g of IPTG and dissolve in 10 ml of sterile water
- Pass through a 0.22 μm syringe filter
- Store at -20 °C
- Weigh out 2.38 g of IPTG and dissolve in 10 ml of sterile water
- 1 M DTT (Dithiothreitol) stock
- Weigh out 1.5 g DTT and dissolve it in 10 ml of sterile water
- Pass through a 0.22 μm syringe filter
- Store in the dark at -20 °C
- Weigh out 1.5 g DTT and dissolve it in 10 ml of sterile water
- 0.5 M EDTA (Disodium Ethylene Diamine Tetra-Acetate) stock (pH 8.0)
- Weigh out 18.16 g of Na2EDTA·2H2O and dissolve in 80 ml of demi-water*
- Adjust to pH 8.0 with pellets of NaOH (~2 g of NaOH is required)
- Fill up to 100 ml with demi-water
- Sterilize by autoclaving Autoclave at 121 °C for 30 min
- Store at room temperature
*Note: The disodium salt of EDTA will not go into solution until the pH of the solution is adjusted to approximately 8.0 by the addition of NaOH.
- Weigh out 18.16 g of Na2EDTA·2H2O and dissolve in 80 ml of demi-water*
- LB medium (1 L)
- Buffers
- Lysis Buffer (1 L)
- Weigh out 29.22 g NaCl (final 500 mM), 2.42 g Tris (final 20 mM) and 0.68 g imidazole (final 10 mM) and dissolve in 900 ml of demi-water
- Adjust pH to 8.0 using HCl
- Fill up to 1 L with demi-water
- Filter using 0.22 µm membrane filter
- Store at 4 °C
- Weigh out 29.22 g NaCl (final 500 mM), 2.42 g Tris (final 20 mM) and 0.68 g imidazole (final 10 mM) and dissolve in 900 ml of demi-water
- Wash Buffer (1 L)
- Weigh out 29.22 g NaCl (final 500 mM), 2.42 g Tris (final 20 mM) and 1.36 g imidazole (final 20 mM) and dissolve in 900 ml of demi-water
- Adjust pH to 8.0 using HCl
- Fill up to 1 L with demi-water
- Filter using 0.22 µm membrane filter
- Store at 4 °C
- Weigh out 29.22 g NaCl (final 500 mM), 2.42 g Tris (final 20 mM) and 1.36 g imidazole (final 20 mM) and dissolve in 900 ml of demi-water
- Elution Buffer (1 L)
- Weigh out 29.22 g NaCl (final 500 mM), 2.42 g Tris (final 20 mM) and 17 g imidazole (final 250 mM) and dissolve in 900 ml of demi-water
- Adjust pH to 8.0 using HCl
- Fill up to 1 L with demi-water
- Filter using 0.22 µm membrane filter
- Store at 4 °C
- Weigh out 29.22 g NaCl (final 500 mM), 2.42 g Tris (final 20 mM) and 17 g imidazole (final 250 mM) and dissolve in 900 ml of demi-water
- Dialysis Buffer (2 L)
- Weigh out 37.27 g KCl (final 250 mM) and 4.77 g HEPES (final 20 mM) and dissolve in 1,900 ml of demi-water
- Adjust pH to 8.0 using KOH
- Fill up to 2 L with demi-water
- Filter using 0.22 µm membrane filter
- Store at 4 °C
- Weigh out 37.27 g KCl (final 250 mM) and 4.77 g HEPES (final 20 mM) and dissolve in 1,900 ml of demi-water
- Dilution Buffer (200 ml)
- Weigh out 0.24 g HEPES (final 10 mM) and dissolve in 100 ml of demi-water
- Adjust pH to 8.0 using KOH
- Fill up to 200 ml with demi-water
- Filter using 0.22 µm membrane filter
- Store at 4 °C
- Weigh out 0.24 g HEPES (final 10 mM) and dissolve in 100 ml of demi-water
- IEX-A Buffer (1 L)
- Weigh out 11.18 g KCl (final 150 mM) and 4.77 g HEPES (final 20 mM) and dissolve in 900 ml of demi-water
- Adjust pH to 8.0 using KOH
- Fill up to 1 L with demi-water
- Filter using 0.22 µm membrane filter
- Store at 4 °C
- Weigh out 11.18 g KCl (final 150 mM) and 4.77 g HEPES (final 20 mM) and dissolve in 900 ml of demi-water
- IEX-B Buffer (1 L)
- Weigh out 149.10 g KCl (final 2 M) and 4.77 g HEPES (final 20 mM) and dissolve in 900 ml of demi-water
- Adjust pH to 8.0 using KOH
- Fill up to 1 L with demi-water
- Filter using 0.22 µm membrane filter
- Store at 4 °C
- Weigh out 149.10 g KCl (final 2 M) and 4.77 g HEPES (final 20 mM) and dissolve in 900 ml of demi-water
- SEC Buffer (1 L)
- Weigh out 37.27 g KCl (final 500 mM), 4.77 g HEPES (final 20 mM) and dissolve in 900 ml of demi-water. Add 1 ml (final 1 mM) of 1 M DTT stock just before use
- Adjust pH to 8.0 using KOH
- Fill up to 1 L with demi-water
- Filter using 0.22 µm membrane filter
- Store at 4 °C
Note: Setting the pH of the buffers at different temperatures will influence the final pH as the pH is temperature dependent. However, in our experience, the pH of the buffers can be set at 4 °C or at RT without having a significant impact on the purification procedure.
- Weigh out 37.27 g KCl (final 500 mM), 4.77 g HEPES (final 20 mM) and dissolve in 900 ml of demi-water. Add 1 ml (final 1 mM) of 1 M DTT stock just before use
- 10x SDS-PAGE Electrophoresis Running Buffer (1 L)
- Weigh out 30.0 g Tris (final 250 mM), 144 g glycine (final 1920 mM), 10 g SDS [final 1% (w/v)] and dissolve in 900 ml of demi-water
- Fill up to 1 L with demi-water
- Weigh out 30.0 g Tris (final 250 mM), 144 g glycine (final 1920 mM), 10 g SDS [final 1% (w/v)] and dissolve in 900 ml of demi-water
- 10x Cas9 Nuclease Reaction Buffer (10 ml)
- Weight out 0.58 g NaCl (final 1 M), 0.1 g MgCl2·6H2O (final 50 mM), 0.476 HEPES (final 200 mM) and 3.72 mg EDTA (final 1 mM) and dissolve in 8 ml of nuclease-free water
- Adjust pH to 6.5 using HCl and fill up to 10 ml with nuclease-free water
- Weight out 0.58 g NaCl (final 1 M), 0.1 g MgCl2·6H2O (final 50 mM), 0.476 HEPES (final 200 mM) and 3.72 mg EDTA (final 1 mM) and dissolve in 8 ml of nuclease-free water
- Lysis Buffer (1 L)
Acknowledgments
This protocol is adapted from Swarts et al. (2017; Reference 26). This work was financially supported by the grant from the Netherlands Organization of Scientific Research (NWO) to J.v.d.O (NWO-TOP, 714.015.001.), by a Swiss National Science Foundation (SNSF) Project Grant to M.J. (SNSF 31003A_149393), and by fellowships of the European Molecular Biology Organization (EMBO) to D.C.S. (EMBO ALTF 179-2015 and EMBO aALTF 509-2017). The authors declare no conflicts of interest or competing interests.
References
- Charpentier, E. and Marraffini, L. A. (2014). Harnessing CRISPR-Cas9 immunity for genetic engineering. Curr Opin Microbiol 19: 114-119.
- Deltcheva, E., Chylinski, K., Sharma, C. M., Gonzales, K., Chao, Y., Pirzada, Z. A., Eckert, M. R., Vogel, J. and Charpentier, E. (2011). CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471(7340): 602-607.
- Doudna, J. A. and Charpentier, E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346(6213): 1258096.
- Endo, A., Masafumi, M., Kaya, H. and Toki, S. (2016). Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida. Sci Rep 6: 38169.
- Ferenczi, A., Pyott, D. E., Xipnitou, A. and Molnar, A. (2017). Efficient targeted DNA editing and replacement in Chlamydomonas reinhardtii using Cpf1 ribonucleoproteins and single-stranded DNA. Proc Natl Acad Sci U S A 114(51): 13567-13572.
- Hsu, P. D., Lander, E. S. and Zhang, F. (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell 157: 1262-1278.
- Hur, J. K., Kim, K., Been, K. W., Baek, G., Ye, S., Hur, J. W., Ryu, S. M., Lee, Y. S. and Kim, J. S. (2016). Targeted mutagenesis in mice by electroporation of Cpf1 ribonucleoproteins. Nat Biotechnol 34(8): 807-808.
- Hu, X., Wang, C., Liu, Q., Fu, Y. and Wang, K. (2017). Targeted mutagenesis in rice using CRISPR-Cpf1 system. J Genet Genomics 44(1): 71-73.
- Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A. and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337(6096): 816-821.
- Karvelis, T., Gasiunas, G., Miksys, A., Barrangou, R., Horvath, P. and Siksnys, V. (2013). crRNA and tracrRNA guide Cas9-mediated DNA interference in Streptococcus thermophilus. RNA Biol 10(5): 841-851.
- Kim, D., Kim, J., Hur, J. K., Been, K. W., Yoon, S. H. and Kim, J. S. (2016a). Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotechnol 34(8): 863-868.
- Kim, H., Kim, S. T., Ryu, J., Kang, B. C., Kim, J. S. and Kim, S. G. (2017). CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat Commun 8: 14406.
- Kim, Y., Cheong, S. A., Lee, J. G., Lee, S. W., Lee, M. S., Baek, I. J. and Sung, Y. H. (2016b). Generation of knockout mice by Cpf1-mediated gene targeting. Nat Biotechnol 34(8): 808-810.
- Kleinstiver, B. P., Tsai, S. Q., Prew, M. S., Nguyen, N. T., Welch, M. M., Lopez, J. M., McCaw, Z. R., Aryee, M. J. and Joung, J. K. (2016). Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat Biotechnol 34(8): 869-874.
- Lei, C., Li, S. Y., Liu, J. K., Zheng, X., Zhao, G. P. and Wang, J. (2017). The CCTL (Cpf1-assisted Cutting and Taq DNA ligase-assisted Ligation) method for efficient editing of large DNA constructs in vitro. Nucleic Acids Res 45(9): e74.
- Li, S. Y., Zhao, G. P. and Wang, J. (2016). C-Brick: A new standard for assembly of biological parts using Cpf1. ACS Synth Biol 5(12): 1383-1388.
- Makarova, K. S. and Koonin, E. V. (2015). Annotation and Classification of CRISPR-Cas Systems. Methods Mol Biol 1311: 47-75.
- Mali, P., Esvelt, K.M. and Church, G. M. (2013). Cas9 as a versatile tool for engineering biology. Nat Methods 10: 957-963.
- Marraffini, L. A. (2015). CRISPR-Cas immunity in prokaryotes. Nature 526(7571): 55-61.
- Mohanraju, P., Makarova, K. S., Zetsche, B., Zhang, F., Koonin, E. V. and van der Oost, J. (2016). Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science 353(6299): aad5147.
- Moreno-Mateos, M. A., Fernandez, J. P., Rouet, R., Vejnar, C. E., Lane, M. A., Mis, E., Khokha, M. K., Doudna, J. A. and Giraldez, A. J. (2017). CRISPR-Cpf1 mediates efficient homology-directed repair and temperature-controlled genome editing. Nat Commun 8(1): 2024.
- Rosenberg, A. H., Lade, B. N., Chui, D. S., Lin, S. W., Dunn, J. J. and Studier, F. W. (1987). Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene 56(1): 125-135.
- Shmakov, S., Smargon, A., Scott, D., Cox, D., Pyzocha, N., Yan, W., Abudayyeh, O. O., Gootenberg, J. S., Makarova, K. S., Wolf, Y. I., Severinov, K., Zhang, F. and Koonin, E. V. (2017). Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol 15(3): 169-182.
- Studier, F. W. and Moffatt, B. A. (1986). Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189: 113-130.
- Studier, F. W., Rosenberg, A. H., Dunn, J. J. and Dubendorff, J. W. (1990). Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol 185, 60-89.
- Swarts, D. C., van der Oost, J. and Jinek, M. (2017). Structural basis for guide RNA processing and seed-dependent DNA targeting by CRISPR-Cas12a. Mol Cell 66(2): 221-233 e224.
- Swiat, M. A., Dashko, S., den Ridder, M., Wijsman, M., van der Oost, J., Daran, J. M., Daran-Lapujade, P. (2017). FnCpf1: a novel and efficient genome editing tool for Saccharomyces cerevisiae. Nucleic Acids Res 45: 12585-12598.
- Tang, X., Lowder, L. G., Zhang, T., Malzahn, A. A., Zheng, X., Voytas, D. F., Zhong, Z., Chen, Y., Ren, Q., Li, Q., Kirkland, E. R., Zhang, Y. and Qi, Y. (2017). A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat Plants 3: 17018.
- van der Oost, J., Westra, E. R., Jackson, R. N. and Wiedenheft, B. (2014). Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol 12(7): 479-492.
- Verwaal, R., Buiting-Wiessenhaan, N., Dalhuijsen, S. and Roubos, J. A. (2017). CRISPR/Cpf1 enables fast and simple genome editing of Saccharomyces cerevisiae. Yeast: 10.1002/yea.3278.
- Wang, M., Mao, Y., Lu, Y., Tao, X. and Zhu, J. K. (2017). Multiplex gene editing in rice using the CRISPR-Cpf1 system. Mol Plant 10(7): 1011-1013.
- Xu, R., Qin, R., Li, H., Li, D., Li, L., Wei, P. and Yang, J. (2017). Generation of targeted mutant rice using a CRISPR-Cpf1 system. Plant Biotechnol J 15(6): 713-717.
- Zaidi, S. S., Mahfouz, M. M. and Mansoor, S. (2017). CRISPR-Cpf1: A new tool for plant genome editing. Trends Plant Sci 22(7): 550-553.
- Zetsche, B., Gootenberg, J. S., Abudayyeh, O. O., Slaymaker, I. M., Makarova, K. S., Essletzbichler, P., Volz, S. E., Joung, J., van der Oost, J., Regev, A., Koonin, E. V. and Zhang, F. (2015). Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163: 759-771.
- Zetsche, B., Heidenreich, M., Mohanraju, P., Fedorova, I., Kneppers, J., DeGennaro, E. M., Winblad, N., Choudhury, S. R., Abudayyeh, O. O., Gootenberg, J. S., Wu, W. Y., Scott, D. A., Severinov, K., van der Oost, J. and Zhang, F. (2017). Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat Biotechnol 35(1): 31-34.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Mohanraju, P., Oost, J. V. D., Jinek, M. and Swarts, D. C. (2018). Heterologous Expression and Purification of the CRISPR-Cas12a/Cpf1 Protein. Bio-protocol 8(9): e2842. DOI: 10.21769/BioProtoc.2842.
Category
Microbiology > Heterologous expression system > Escherichia coli
Microbiology > Microbial biochemistry > Protein > Activity
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











