- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Method for Extracting the Nuclear Scaffold from the Chromatin Network
Published: Vol 8, Iss 8, Apr 20, 2018 DOI: 10.21769/BioProtoc.2821 Views: 7594
Reviewed by: Manjula MummadisettiPearl CampbellAmey Redkar

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Modified Pseudo-Schiff Propidium Iodide for Staining the Shoot Apical Meristem in Arabidopsis
Ruiqi Li [...] Ligeng Ma
May 5, 2023 2269 Views

Dissecting the Mechanical Control of Mitotic Entry Using a Cell Confinement Setup
Margarida Dantas [...] Jorge G. Ferreira
Mar 20, 2024 2732 Views
Abstract
Each cell contains many large DNA polymers packed in a nucleus of approx. 10 μm in diameter. With histones, these DNA polymers are known to form chromatins. How chromatins further compact in the nucleus is unclear but it inevitably depends on an extensive non-chromatin nuclear scaffold. Imaging of endogenous chromatin network and the complementary scaffold that support this network has not been achieved but biochemical and proteomic investigations of the scaffold can still provide important insights into this chromatin-organizing network. However, this demands highly inclusive and reproducible extraction of the nuclear scaffold. We have recently developed a simple protocol for releasing the scaffold components from chromatins. The inclusiveness of the extract was testified by the observation that, upon its extraction from the nuclei, the remaining nuclear chromatins were liberated into extended and often parallel chromatin fibers. Basically, this protocol includes the generation of pure nuclei, treatment of the nuclei with Triton X-100 to generate envelope-depleted nuclei (TxN), and extraction of the nuclei at 500 mM NaCl in a sucrose-containing buffer. This combined extract of TxN is known as TxNE.
Keywords: NucleiBackground
Chromatins are densely and dynamically compacted in the nuclei through a complex scaffold of proteins and ribonucleoproteins. Unlike the cytoskeletal networks (Fischer and Fowler, 2015), microscopic observation of this nuclear scaffold is technically challenging. This may reflect the dominance of chromatins inside each nucleus with which the scaffold mingles and negotiates in the nucleus. The spherical arrangement of the nucleus also contributes to challenges in imaging such scaffold structures. A major element of the nuclear scaffold is the nuclear lamina (NL) (Gruenbaum and Foisner, 2015). NL covers the surface of the entire nuclear chromatin mass on which the nuclear envelop (NE) attaches. The peripheral surface of the nuclear chromatin mass is characterized by dense heterochromatin (Gruenbaum and Foisner, 2015). Inside the nucleus, one or more nucleoli can be found that occupy significant nuclear regions (Jordan and McGovern, 1981; Shaw and Jordan, 1995; Pederson, 2011). The nucleolar surface is also surrounded by a rim of dense chromatins and potentially functions as an important interior scaffold to support the chromatins (Chen et al., 2018). The morphology of the nucleoli changes dynamically depending on the stage of the cell cycle which can significantly affect the organization of nuclear chromatins (Hernandez-Verdun, 2011; Chen et al., 2018).
The presence of such scaffold structures has been extensively investigated in 1980-90s although a direct visualization has not been achieved (Gasser, 2002; Laemmli et al., 1992). However, the composition of the nuclear scaffold and interactions among the scaffold elements could be learned through biochemical, proteomic and imaging studies. To this end, effective and reproducible extraction of the nuclear scaffold from the chromatin network is essential. We have recently developed a simple protocol for the extraction of nuclear scaffold proteins (Chen et al., 2018).
This protocol was initially developed based on a study showing that isolated nucleoli could be solubilized at 400 mM NaCl (Trimbur and Walsh, 1993). We adopted this condition to solubilize isolated nuclei. The inclusiveness of the extract was testified by the liberation of nuclear chromatins as extended parallel chromatin fibers (Chen et al., 2018). TxNE can be used to study many aspects of the nuclear scaffold.
Materials and Reagents
- 0.5-10 L pipette tips (Corning, Axygen®, catalog number: T-300 )
- 10-200 ml pipette tips (Greiner Bio One International, catalog number: 739290 )
- 100-1,000 ml pipette tip (Greiner Bio One International, catalog number: 686290 )
- T 175 cell culture flask (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 159910 )
- T 75 cell culture flasks (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 156499 )
- 150 mm circular tissue culture dishes (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 168381 )
- 50 ml Falcon tubes (Greiner Bio One International, catalog number: 227261 )
- 5 ml blunt-ended syringes (5 ml/cc, Luer slip, Cellotron, https://dcellotron.en.ec21.com/)
- 15 ml Falcon tubes (Greiner Bio One International, catalog number: 188271 )
- Nalgene Oak Ridge High-Speed Centrifuge Tubes (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 3114-0030 )
- Regular 1.5 ml microcentrifuge tubes (Corning, Axygen®, catalog number: MCT-150-C )
- Low adhesion microcentrifuge tubes (Eppendorf, catalog number: 022431081 )
- 12 mm glass coverslips (Electron Microscopy Sciences, catalog number: 72231-10 )
- Glass slides (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 6776215 )
- 50 ml syringes (Terumo, catalog number: SS*50LE )
- 0.22 μm filters (Merck, catalog number: SLGP033RS )
- HeLa S3 human cervical carcinoma cells (ATCC, catalog number: CCL-2 )
- Trypsin solution (0.05% w/v, Capricorn scientific, catalog number: TRY-1B10 )
- 0.5% Trypsin (Trypsin-EDTA, 10x stock, Thermo Fisher Scientific, GibcoTM, catalog number: 15400054 )
- Protease inhibitors PMSF (BDH, catalog number: 442172C )
- Triton X-100 (Sigma-Aldrich, catalog number: T8787 )
- 5 M NaCl
- 1% (w/v) paraformaldehyde
- Mouse anti-nucleophosmin-1 (NPM1) antibody (Sigma-Aldrich, catalog number: B0556 )
- Rabbit anti-lamin B1 (LB1) antibody (Abcam, catalog number: ab16048 )
- Rabbit anti-histone H1.2 (Abcam, catalog number: ab17677 )
- Rabbit anti-lamin A/C (Santa Cruz Biotechnology, catalog number: sc-7292 )
- Rabbit anti-H1.x antibody (Abcam, catalog number: ab31972 )
- Goat anti-mouse IgG (Cy3-conjugated) (Jackson ImmunoResearch Laboratories, catalog number: 115-165-003 )
- Goat anti-rabbit IgG (Alexa Fluor 488-conjugated) (Jackson ImmunoResearch Laboratories, catalog number: 115-545-003 )
- Mounting medium (Vector laboratories, catalog number: H-1200 )
- Any clear Nail polish (We use Daiso Japan Nail Top Coat Clear, 10 ml bottle, made in Taiwan)
- 12.5% (w/v) gels
- Sodium chloride (NaCl) (Merck)
- Potassium chloride GR (KCl)(Merck)
- Di-sodium hydrogen phosphate (Merck)
- Sucrose (First BASE Laboratories, catalog number: BIO-1090-1kg )
- Sucrose (Merck)
- Tris (Merck)
- Tris base (Avantor Performance Materials, J.T. Baker, catalog number: 4109-02 )
- Magnesium chloride hexahydrate (Merck, catalog number: 1058331000 )
- Magnesium chloride hexahydrate (MgCl2·H2O)( Merck)
- DMEM medium (Thermo Fisher Scientific, GibcoTM, catalog number: 12430054 )
- Fetal bovine serum (FBS, GE Healthcare, HyClone, catalog number: SV30160.03 )
Note: We purchased FBS and generated heat-inactivated FBS by incubation at 56 °C for 30 min. - Penicillin-Streptomycin (100x stock, PAN Biotech, catalog number: P06-07100 )
- L-glutamine (200 mM, 100x stock, Thermo Fisher Scientific, GibcoTM, catalog number: 25030081 )
- PBS, pH 7.4 (see Recipes)
- 0.25 M sucrose buffer, pH 7.4 (see Recipes)
- 2.2 M sucrose buffer, pH 7.4 (see Recipes)
- DMEM culture medium (see Recipes)
Equipment
- 1,000 L pipette (Eppendorf, model: Research® plus, catalog number: 3120000062 )
- 200 L pipette (Eppendorf, model: Research® plus, catalog number: 3120000054 )
- Humidified cell incubator (Thermo Fisher Scientific, Thermo ScientificTM, model: FormaTM Steri-CycleTM CO2 incubator , catalog number: 371)
- Homogenizer (Isobiotec Precision Engineering, Heidelberg, Germany)
- Tabletop microcentrifuge (Beckman Coulter, model: Microfuge 22R centrifuge , catalog number: 368831)
- Tabletop swing bucket centrifuge (Eppendorf, model: 5810 R , catalog number: 5811000320)
- Beckman Coulter centrifuge with a JA-20 rotor (Beckman Coulter, model: Avanti J-25 , catalog number: 363102)
- BSL-2 cell culture cabinet (Gelman, model: Bioessential Class II series Type A2 Laminar flow BSC, http://gelmansingapore.com/product/bioessential-class-ii-type-a2-series-laminar-flow-biological-safety-cabinets/)
- FluoView FV1000 confocal microscope
- Cool/SNAP HQ2 image acquisition camera (Olympus)
Software
- FV-ASW 1.6b software
- Imaris software (Bitplane AG)
Procedure
- Generation of cell homogenate
- Culture HeLa cells in a T-75 flask in a humidified BSL-2 incubator at 37 °C containing 25 ml of DMEM culture medium to confluence.
- Divide the cells equally five 150 mm round culture dishes and culture in the DMEM culture medium (25 ml/plate) to confluence.
- Remove the media and wash each plate using 10 ml of PBS.
- Add 2 ml of 1x trypsin solution to cover the surface of each plate and incubate the plates for 2 min at 37 °C in a humidified cell incubator.
- Tap the plates horizontally to suspend the cells and add 6 ml of culture medium to each plate.
- Harvest cells from each plate and combine in a 50 ml Falcon tube.
- Harvest cells by centrifugation for 5 min at 400 x g.
- Re-suspend cells in 50 ml of PBS and wash by centrifugation for 5 min at 400 x g. Repeat this step once.
- Re-suspend cells in 4 ml of 0.25 M sucrose buffer containing the protease inhibitor PMSF (0.25 mM) and pipet to get a single cell suspension.
- Assemble an isobiotec cell homogenizer using a 10 μm metal ball and pre-chill the cell homogenizer on ice (Figure 1).
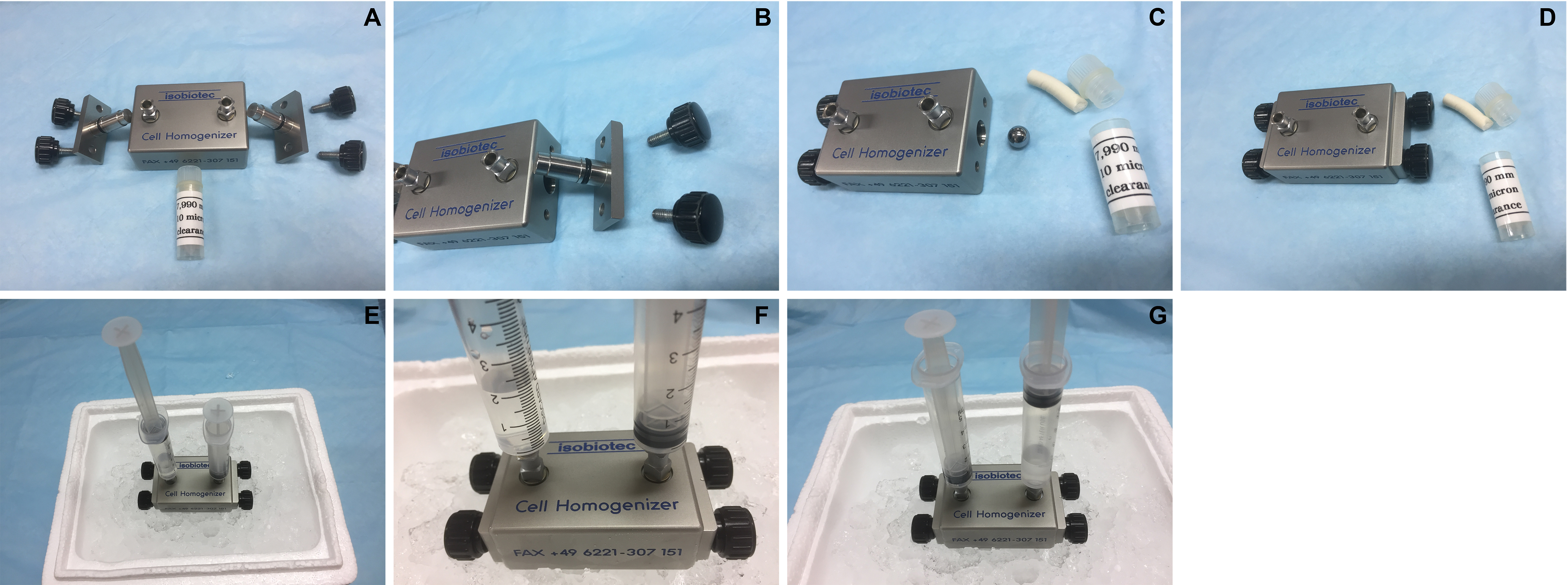
Figure 1. Assembly of the Isobiotec cell homogenizer. A. Parts of the homogenizer including the metal ball (10 μm clearance) in the white tube; B. Side view; C. The metal ball is placed in the central passage. D. Completed assembly of the homogenizer. E. Each outlet is fitted with a 5 ml syringe. F. 2 ml of cell suspension is added to one syringe (water is used in this demonstration). G. Press to pass the cell suspension through the metal ball-fitted central passage into the other syringe. Repeat another 11 times before harvesting the cell homogenate. - Fit the two outlets on the homogenizer each with a 5 ml syringe.
- Remove the plunger from one syringe to add 2 ml of the cell suspension.
- Pass the cell suspension 12 times through the homogenizer.
- Transfer the homogenized cell lysate into a 15 ml Falcon tube on ice and repeat homogenization with the rest of the cell suspension.
- Wash the homogenizer 2 times each with 1 ml of 0.25 M sucrose buffer and combine with the lysate.
- Culture HeLa cells in a T-75 flask in a humidified BSL-2 incubator at 37 °C containing 25 ml of DMEM culture medium to confluence.
- Isolation of nuclei
- Centrifuge the cell homogenate for 10 min at 600 x g and 4 °C to pellet the nuclei.
- Re-suspend the pellet in 5 ml of 2.2 M sucrose buffer in Nalgene Oak Ridge High-Speed Centrifuge Tubes.
- Centrifuge for 30 min at 50,000 x g and 4 °C.
- Remove supernatant and add 2 ml of 0.25 M sucrose buffer to rinse the tube area near the pellet.
- Remove the sucrose buffer without disturbing the pellet.
- Re-suspend the pellet in 4 ml of 0.25 M sucrose buffer and transfer to a 15 ml Falcon tube.
- Centrifuge for 5 min at 600 x g and 4 °C.
- Centrifuge the cell homogenate for 10 min at 600 x g and 4 °C to pellet the nuclei.
- Depletion of the nuclear envelope (NE)
- Preparation of Triton-containing sucrose buffer.
- Add 0.1 ml of Triton X-100 in a 15 ml tube along the tube wall.
- Add 10 ml of 0.25 M sucrose buffer to the tube and vortex till Triton is mixed thoroughly with the sucrose buffer.
- Add 0.1 ml of Triton X-100 in a 15 ml tube along the tube wall.
- Re-suspend the nuclear pellet in 4 ml of Triton-containing sucrose buffer and incubate for 1 h on ice.
- Centrifuge for 5 min at 600 x g and 4 °C.
- Re-suspend the pellet in another 5 ml of Triton-containing sucrose buffer.
- Centrifuge for 5 min at 600 x g and 4 °C.
- Wash these Triton-extracted nuclei (TxN) 2 times using 0.25 M sucrose buffer.
- Then resuspend the pellet in 0.9 ml of 0.25 M sucrose buffer.
- Preparation of Triton-containing sucrose buffer.
- Extraction of the nuclear scaffold (Figure 2)
- Add 0.1 ml 5 M NaCl solution to the nuclear suspension (colloidal gel forms immediately at the interface).
- Pipette vigorously 20-30 times using a 1,000 L Eppendorf pipette until the entire suspension turns into a uniform colloidal gel.
- Add 0.5 ml of 0.25 M sucrose buffer containing 0.5 M NaCl and pipette to homogeneity.
- Centrifuge for 10 min at 2,000 x g and recover a small volume of supernatant if it is clearly observed.
- Add 0.5 ml of 0.25 M sucrose buffer and the colloidal gel should shrink immediately.
- Poke the gel vigorously using a 0.2 ml pipette tip so that the gel continues to shrink.
- Centrifuge for 10 min at 2,000 x g and 4 °C and recover the clear supernatant.
- Add another 0.5 ml of 0.25 M sucrose buffer to the pellet and continue to poke the fragment until it ceases shrinking (It should look like a brown and non-transparent fragment).
- Centrifuge again and combine all the supernatants as the extract.
- Measure the A280 absorption of the combined TxN extracts or TxNE. It should range 1.0-2.0.
- Aliquot TxNE in non-adhesive 1.5-ml microtubes and store at -80 °C.
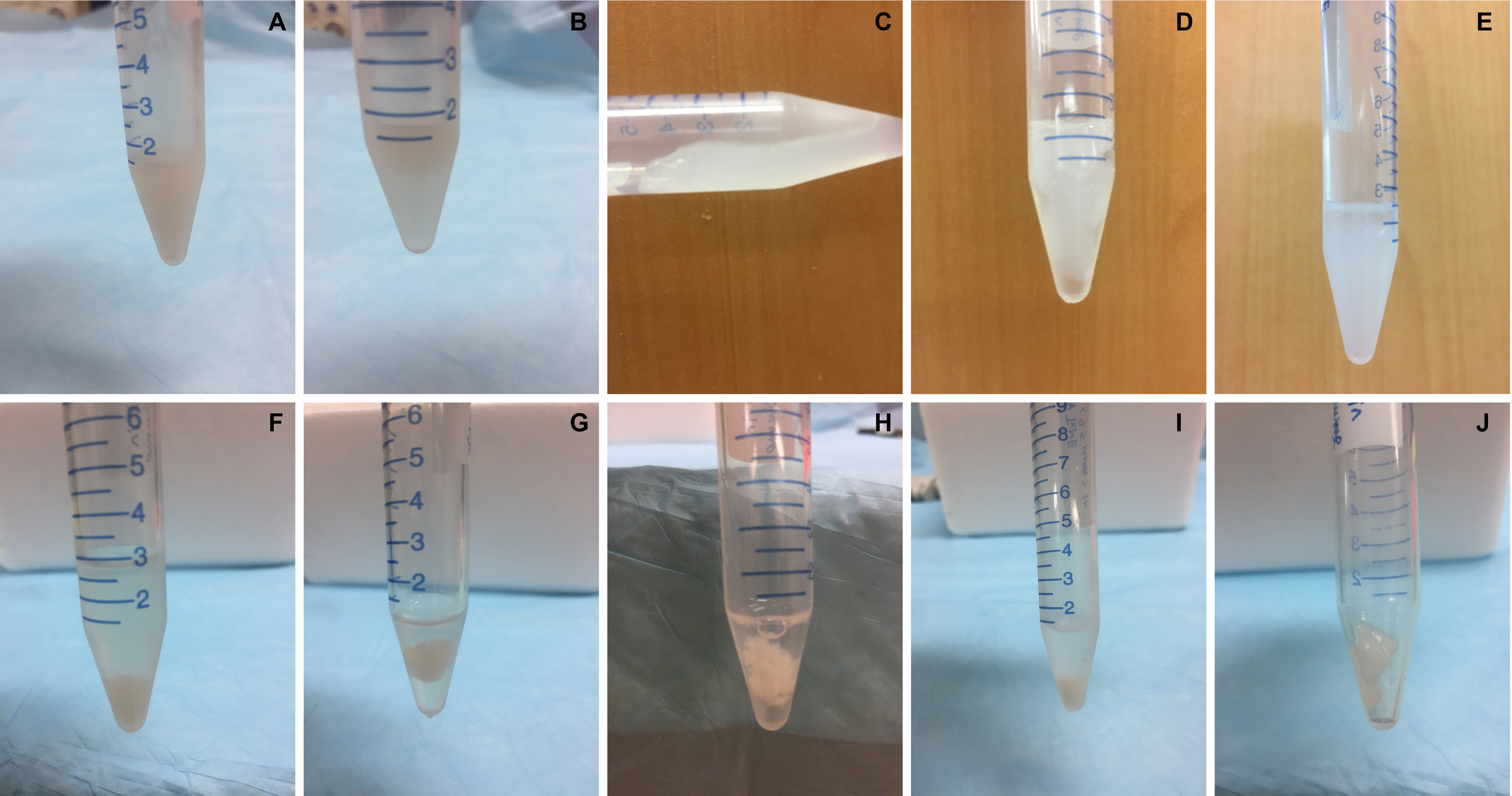
Figure 2. Extraction of TxNE from the nuclei. A. Single cell suspension generated from 6 x 108 HeLa cells in 1.35 ml of 2.5 M sucrose buffer. B. After adding 0.15 ml of 5 M NaCl, the interphase moved rapidly upward giving a transparent lower phase in less than 30 sec. C. After vigorously pipetting for ~30 times in less than 1 min, the entire cell suspension became one single colloidal gel. D. 0.75 ml of the 0.25 M sucrose buffer containing 0.5 M NaCl was added to the gel and mixed by tapping for ~20 times. E. Centrifuge for 10 min at 2,000 x g and ~0.2 ml supernatant can be recovered from the top. F. Add 0.75 ml of the 0.25 sucrose buffer (without additional NaCl) and mix vigorously for 1-2 min to observe the gel shrinking. Poke the gel 20-50 times with a 10-200 L tip fitted on a pipette until the gel stop shrinking. Centrifuge for 10 min at 2,000 x g and harvest the supernatant. G. Add 0.75 ml of the 0.25 M sucrose buffer to the pellet. H. Continue to ~50 times until when the pellet shrunk into an irregular fragment. Some smaller fragments are also visible. I. Centrifuge for 10 min at 1,000 x g to recover the supernatant. J. The supernatants are combined as TxNE and the pellet is typically the extracted chromatins which retained the cellular DNA and core histones.
Note: To show the sample status in each step more clearly, we have scaled up the solution volumes 1.5x of that described in the main text of the protocol for photograph.
- Add 0.1 ml 5 M NaCl solution to the nuclear suspension (colloidal gel forms immediately at the interface).
Data analysis
- The usual protein profile of TxNE is highly consistent (Chen et al. 2018). During TxNE isolation, some steps can be monitored for consistency. Firstly, the nuclei isolated after centrifugation through 2.2 M sucrose can be examined by confocal microscopy by immuno-staining of the signature NL protein lamin B1 and the signature nucleolar protein nucleophosmin-1 (NPM1) (Figure 3). The isolated nuclei mostly remain continuous in the LB1-defined NL although the nucleoli can sometimes become enlarged. The nuclei mostly remain oval in morphology. These nuclei should be free of vimentin although it often associates significantly with the nuclear surface if the nuclei are not centrifuged through the 2.2 M sucrose buffer. TxN should remain individual although these NE-depleted nuclei tend to form clusters. The LB1-defined NL should be mostly continuous although the shapes of TxN can be less oval in morphology (Figure 3).
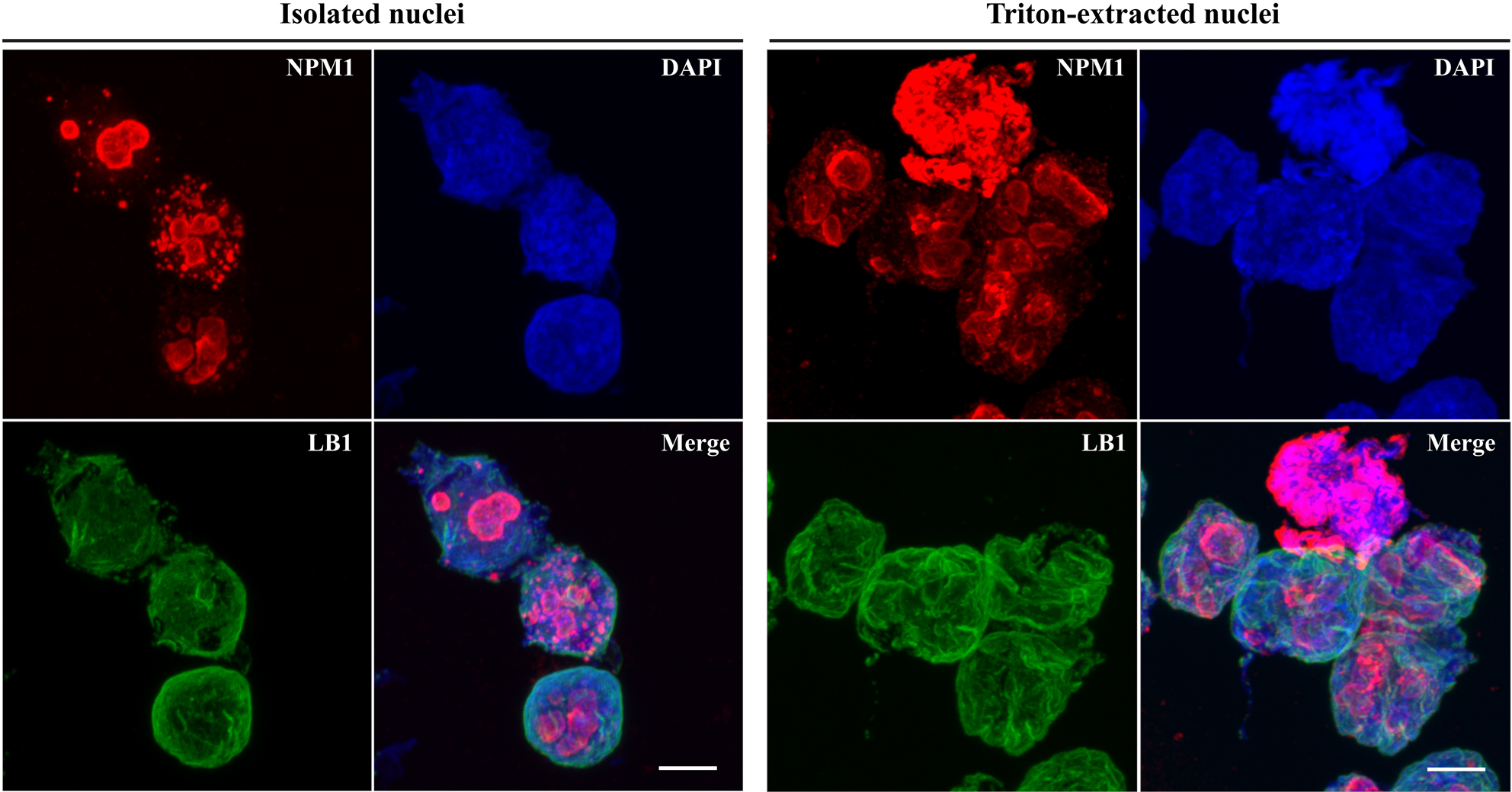
Figure 3. Integrity of isolated and Triton-extracted nuclei. HeLa cells homogenized and nuclei were isolated by centrifugation through the 2.2-M sucrose buffer. The nuclei were incubated with 12-mm glass coverslips for 5 min on ice and then fixed for 30 min in 1% (w/v) paraformaldehyde. The nuclei were then incubated for 30 min in 1% (v/v) Triton X-100 in PBS. The nuclei were also re-suspended in the 0.25-M sucrose buffer containing 1% (v/v) Triton X-100 and incubated for 1 h on ice to generate Triton-extracted nuclei (TxN) (right panel). TxN was washed in 0.25 M sucrose buffer and then incubated for 5 min on ice with 12-mm glass coverslips. The coverslips were fixed for 30 min in 1% (w/v) paraformaldehyde. All coverslips were first incubated for 30 min with mouse anti-NPM1 and rabbit anti-LB1 antibodies and were, after washing, incubated for 30 min with goat anti-rabbit (Alexa Fluor 488, green) and anti-mouse (Cy3, red) IgG. After washing, coverslips were mounted using DAPI-containing VectorShield medium and viewed using a FluoView FV1000 confocal microscope equipped with a 100x oil objective (aperture 1.45) and Cool/SNAP HQ2 image acquisition camera (Olympus). Images are captured with the FV-ASW 1.6b software and analyzed using the Imaris software (Bitplane AG). Scale bars = 5 μm. - TxNE does not contain noticeable levels of core histones (Chen et al., 2018). However, the linker H1 histone is included in this extract (Figure 4). Little LB1 is extracted in TxNE (Figure 4). LB1 remains associated with the extracted chromatins although it is dispersed among the extended chromatins as fragments (Chen et al., 2018). In contrast, lamin A/C is effectively extracted with TxNE (Figure 4).
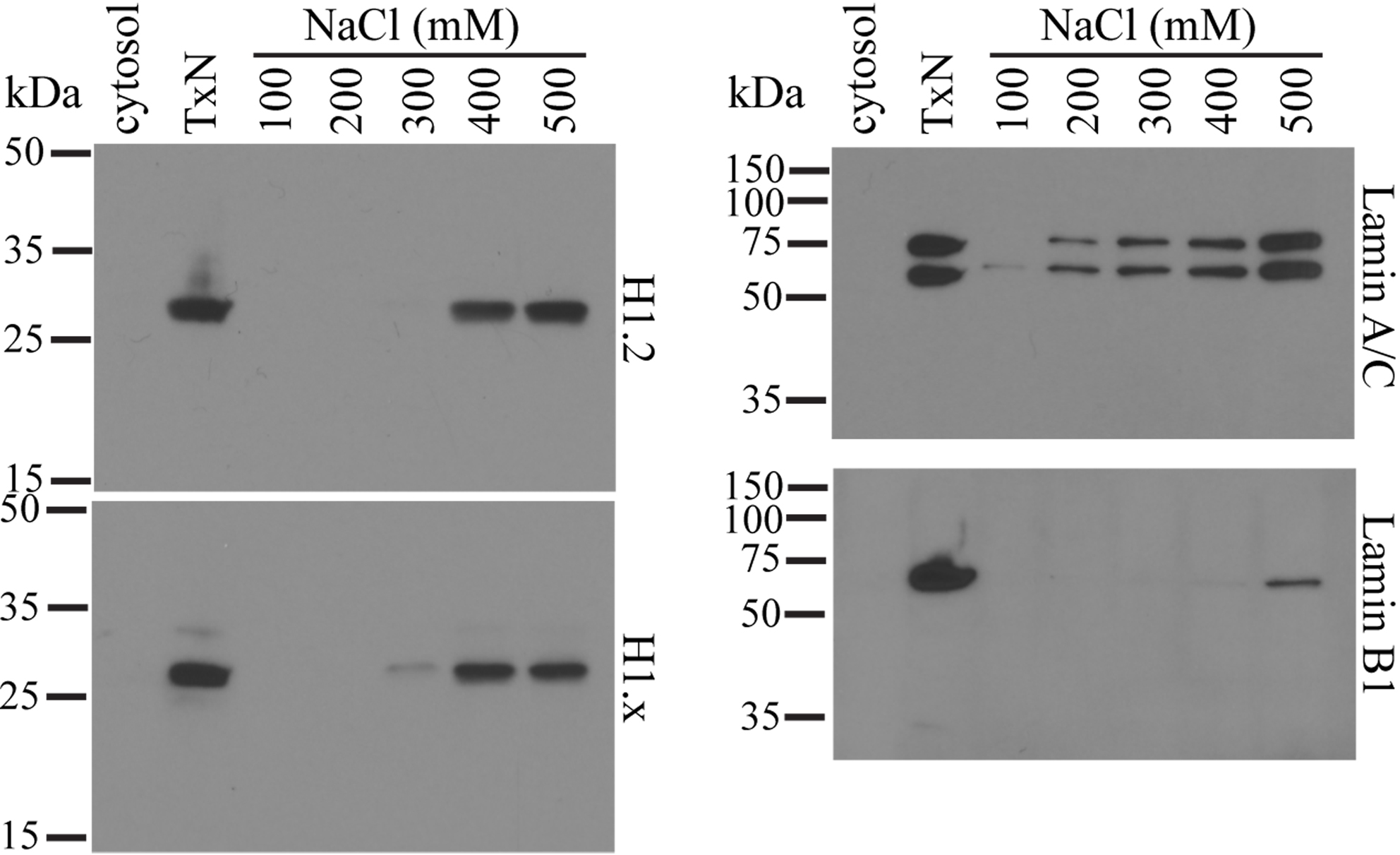
Figure 4. Selective elution of nuclear proteins from the chromatin network. Nuclei are isolated and depleted of NE with Triton X-100. These NE-depleted nuclei (TxN) were extracted with 0.25 M sucrose buffer containing 100, 200, 300, 400 or 500 mM of NaCl. Equal volumes of the supernatants were subjected to SDS-PAGE on 12.5% (w/v) gels and analyzed by Western blotting. The cytosol fraction and TxN used in the extraction were also included as controls. All antibodies used are affinity-purified polyclonal rabbit antibodies obtained from Abcam.
Notes
- This protocol is highly reproducible as stated. However, researchers may encounter difficulties during cell homogenization if single cell suspension is not achieved.
- This protocol is optimized for HeLa cells. For other cell types, the metal ball diameter needs to be optimized. The quality of nuclei thus generated can be assessed by simple immuno-staining for LB1, chromatins, and NPM1.
- TxNE should be examined by SDS-PAGE and Coomassie Blue staining to assess consistency in protein profiles (Chen et al., 2018).
Recipes
- PBS (pH 7.4)
137 mM NaCl
2.7 mM KCl
10 mM disodium hydrogen phosphate - 0.25 M sucrose buffer (pH 7.4)
0.25 M sucrose
10 mM Tris
5.0 mM MgCl2
Filter through a 0.22 μm filter and store at 4 °C - 2.2 M sucrose buffer (pH 7.4)
2.2 M sucrose
10 mM Tris
5.0 mM MgCl2
Filter through a 0.22 μm filter and store at 4 °C - DMEM culture medium
450 ml of DMEM medium
50 ml of fetal bovine serum (FBS)
5 ml of penicillin-streptomycin (100x stock)
5 ml of L-glutamine (200 mM, 100x stock)
Acknowledgments
The authors thank Shu Ying Lee for assistance in confocal microscopy. This project was supported by a Singapore National University Health System Basic Research grant (NUHSRO/2011/005/STB/B2B-01) and a National Medical Research Council grant (NMRC/OFIRG/0013/2016). Authors declare no conflict of interest or competing interests.
References
- Chen, J., Teo, B. H. D., Cai, Y., Wee, S. Y. K. and Lu, J. (2018). The linker histone H1.2 is a novel component of the nucleolar organizer regions. J Biol Chem 293(7): 2358-2369.
- Fischer, R. S. and Fowler, V. M. (2015). Thematic minireview series: The state of the cytoskeleton in 2015. J Biol Chem 290(28): 17133-17136.
- Gasser, S. M. (2002). Visualizing chromatin dynamics in interphase nuclei. Science 296(5572): 1412-1416.
- Gruenbaum, Y., and Foisner, R. (2015). Lamins: nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu Rev Biochem 84: 131-164.
- Hernandez-Verdun, D. (2011). Assembly and disassembly of the nucleolus during the cell cycle. Nucleus 2(3): 189-194.
- Jordan, E. G. and McGovern, J. H. (1981). The quantitative relationship of the fibrillar centres and other nucleolar components to changes in growth conditions, serum deprivation and low doses of actinomycin D in cultured diploid human fibroblasts (strain MRC-5). J Cell Sci 52: 373-389.
- Laemmli, U. K., Kas, E., Poljak, L. and Adachi, Y. (1992). Scaffold-associated regions: cis-acting determinants of chromatin structural loops and functional domains. Curr Opin Genet Dev 2(2): 275-285.
- Pederson, T. (2011). The nucleolus. Cold Spring Harb Perspect Biol 3(3).
- Shaw, P. J. and Jordan, E. G. (1995). The nucleolus. Annu Rev Cell Dev Biol 11: 93-121.
- Trimbur, G. M. and Walsh, C. J. (1993). Nucleolus-like morphology produced during the in vitro reassociation of nucleolar components. J Cell Biol 122(4): 753-766.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Chen, J., Teo, B. H. D. and Lu, J. (2018). A Method for Extracting the Nuclear Scaffold from the Chromatin Network. Bio-protocol 8(8): e2821. DOI: 10.21769/BioProtoc.2821.
- Chen, J., Teo, B. H. D., Cai, Y., Wee, S. Y. K. and Lu, J. (2018). The linker histone H1.2 is a novel component of the nucleolar organizer regions. J Biol Chem 293(7): 2358-2369.
Category
Cell Biology > Cell structure > Nucleus
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









