- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantification of Colibactin-associated Genotoxicity in HeLa Cells by In Cell Western (ICW) Using γ-H2AX as a Marker
Published: Vol 8, Iss 6, Mar 20, 2018 DOI: 10.21769/BioProtoc.2771 Views: 8073
Reviewed by: Andrea PuharIsabel SilvaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Adhesion of Enteroaggregative E. coli Strains to HEK293 Cells
Jorge Luis Ayala-Lujan and Fernando Ruiz-Perez
Apr 20, 2018 6850 Views

Intracellular Invasion and Killing Assay to Investigate the Effects of Binge Alcohol Toxicity in Murine Alveolar Macrophages
Victor Jimenez Jr and Fernando P Monroy
Jan 20, 2019 6733 Views

Model of Chemotherapy-associated Mucositis and Oral Opportunistic Infections
Takanori Sobue [...] Anna Dongari-Bagtzoglou
Nov 5, 2019 5322 Views
Abstract
The genotoxin colibactin is produced by several species of Enterobacteriaceae. This genotoxin induces DNA damage, cell cycle arrest, senescence and death in eukaryotic cells (Nougayrède et al., 2006; Taieb et al., 2016). Here we describe a method to quantify the genotoxicity of bacteria producing colibactin following a short infection of cultured mammalian cells with colibactin producing E. coli.
Keywords: ColibactinBackground
The genotoxin colibactin is a polyketide nonribosomal peptide hybrid compound produced by several species of Enterobacteriaceae. This toxin, synthesized by a machinery encoded on a 54 kb genomic locus, the pks island, induces DNA damages, cell cycle arrest, senescence and death in eukaryotic cells (Nougayrède et al., 2006; Taieb et al., 2016). The genotoxic activity of colibactin is dependent on a direct host cell-bacteria interaction and cannot be recapitulated from culture supernatant, killed bacteria, or bacterial lysates instead of life bacteria. Visualization and quantification of the colibactin genotoxic effect on eukaryotic cells can be assessed by quantification of the megalocytosis phenotype (for a protocol see Bossuet-Greif et al., 2017) or quantification of the double-strand DNA breaks in the host cell nucleus by a comet assay (revealing DNA fragmentation) or phosphorylation of the H2AX histone, a marker of double-strand DNA breaks. The phosphorylation of the histone H2AX is characterized as an early and sensitive reaction to genotoxic agents (Audebert et al., 2010). H2AX phosphorylation was demonstrated to be 10-100 times more sensitive than the comet assay in vitro as well as in vivo (Audebert et al., 2010). The quantification of phosphorylated histone H2AX (γ-H2AX) can be processed by the In-Cell Western Assay, an immunochemical assay that uses fluorescence to detect and quantify proteins in fixed cells (Audebert et al., 2010). Here we describe an adapted assay allowing the measurement of γ-H2AX in 96-well plate using In-Cell Western, following a short infection of cultured mammalian cells with colibactin-producing bacteria (Martin et al., 2013; Bossuet-Greif et al., 2016; Tronnet et al., 2017).
Materials and Reagents
- Black tissue culture plate 96 wells flat bottom (Greiner Bio One International, catalog number: 655090 )
- Parafilm
- Aluminum foil
- pks+ and pks- Escherichia coli strains (stored in LB 20% glycerol at -80 °C)
Note: Strains typically used as positive controls in the authors’ laboratory are probiotic strain Nissle 1917 or the commensal strain M1/5. Strains used as a negative control are the K-12 strain MG1655. Our lab can provide these strains. - HeLa cells (ATCC, catalog number: CCL-2 ), 20 passages maximum
- Lennox L broth base (LB medium; Thermo Fisher Scientific, InvitrogenTM, catalog number: 12780029 )
- Dulbecco’s modified Eagle medium (DMEM) with 25 mM HEPES (Thermo Fisher Scientific, GibcoTM, catalog number: 42430 )
- Hanks’ balanced salt solution (HBSS; Sigma-Aldrich, catalog number: H8264 )
- Gentamicin solution 50 mg/ml (Sigma-Aldrich, catalog number: G1397 )
- Dulbecco’s phosphate buffered saline (PBS; Sigma-Aldrich, catalog number: D8537 )
- Dulbecco’s modified Eagle medium (DMEM), high glucose, GlutaMax Supplement, pyruvate (Thermo Fisher Scientific, GibcoTM, catalog number: 31966021 )
- Fetal bovine serum (FBS; Thermo Fisher Scientific, GibcoTM, catalog number: 10270106 )
- Non-essential amino acids solution (NEAA) 100x (Thermo Fisher Scientific, GibcoTM, catalog number: 11140035 )
- Paraformaldehyde (PFA) 20% (Electron Microscopy Sciences, catalog number: 15713 )
- 10x PBS (Sigma-Aldrich, catalog number: D1408 )
- Ammonium chloride (NH4Cl; BioUltra, Sigma-Aldrich, catalog number: 09718-250G )
- TritonTM X-100 (Sigma-Aldrich, catalog number: X100-500ML )
- MAXblock blocking medium (Active Motif, catalog number: 15252 )
- Phosphatase inhibitor PHOSTOP (10x, Roche Diagnostics, catalog number: 04906837001 )
- RNase (Sigma-Aldrich, catalog number: R6513 )
- Sodium chloride (NaCl; Sigma-Aldrich, catalog number: S7653 )
- Sodium azide (Sigma-Aldrich, catalog number: S8032 )
- Rabbit monoclonal anti-γ-H2AX antibody (Cell Signaling Technology, catalog number: 9718 )
- IRDyeTM 800CW-conjugated goat anti-rabbit secondary antibody (2 mg/ml, Biotium (distributor Interchim), catalog number: 20078 )
- RedDotTM2 (200x in DMSO, Biotium (distributor Interchim), catalog number: 40061 )
- HeLa cell culture medium (see Recipes)
- Fixation solution (see Recipes)
- Neutralization solution (see Recipes)
- 10% Triton X-100 (see Recipes)
- Permeabilization solution (see Recipes)
- Blocking solution (see Recipes)
- Permeabilization solution (see Recipes)
- PST buffer (see Recipes)
- 100x azide (see Recipes)
- Anti-γ-H2AX solution (see Recipes)
- Secondary antibody solution (see Recipes)
Equipment
- 37 °C, 5% CO2 incubator for cell cultures
- 37 °C incubator for bacterial cultures
- Microplate reader with 680 and 800 nm channels (here, Odyssey Infrared Imaging Scanner, Li-Cor ScienceTec, Les Ulis, France)
- Microplate reader for absorbance measurement at 600 nm (TECAN Infinite Pro)
- Colorimeter to measure the absorbance at 600 nm of bacterial cultures (Biochrom, model: WPA CO7500 )
- Variable Speed Rocker (VWR, catalog number: 75832-308 )
- Chemical safety hood
- Clean bench
- Inverted microscope (Olympus, model: CKX31 )
- Pipettes and multichannel micropipette 30-300 µl
Software
- GraphPad Prism 6.0
Procedure
Day 1
- Bacteria are cultured overnight in 3 ml of LB from -80 °C glycerol stock or a colony on an LB agar plate at 37 °C with shaking (220 rpm).
- HeLa cells are dispensed in a black 96-well tissue culture plate with transparent bottom (1.5 x 104 cells/well) and grown for 24 h in 200 μl HeLa cell culture medium (see Recipes) at 37 °C in a CO2 incubator.
Note: Importantly, don’t dispense any cells in the two bottom-left wells (see Figure 1: plate infection scheme).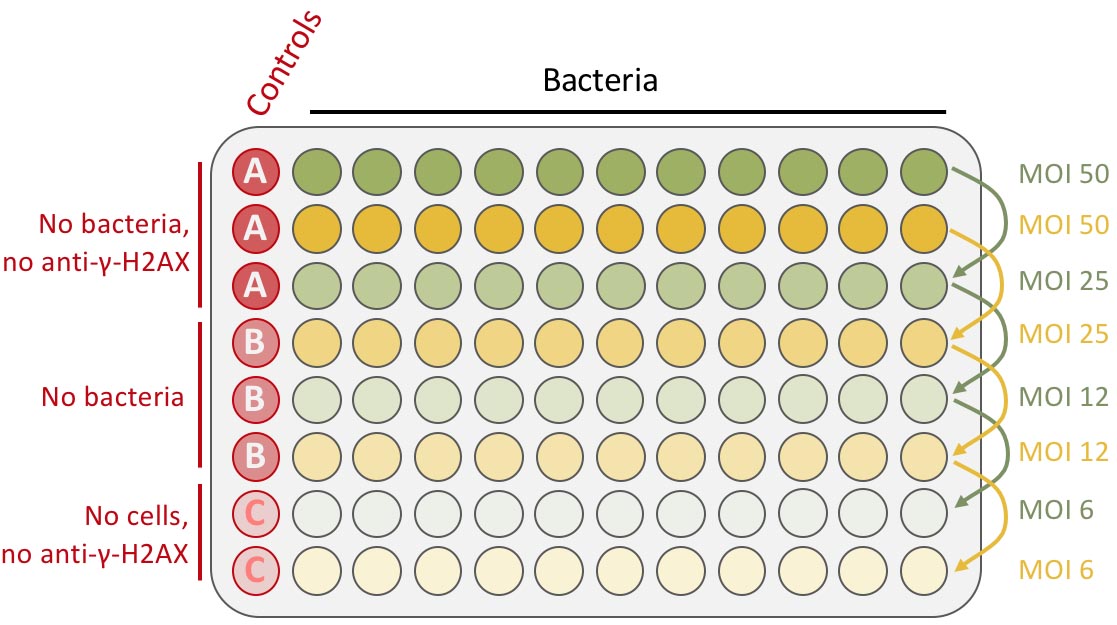
Figure 1. Schematic representation of plate infection example. Bacteria are inoculated in the two first rows in duplicate (in 200 μl/well of DMEM-HEPES medium), at an MOI of 50 and then 2-fold serially diluted (by successively transferring 100 μl to the rows below). The final volume is 100 μl. The left column is kept for the controls (red wells column).
Day 2
- Bacterial preparation
Bacteria are sub-cultured by diluting 50 µl of the overnight culture in 5 ml of DMEM-HEPES medium (i.e., 1:100 dilution) and grown at 37 °C with agitation (220 rpm) until an average OD600nm = 0.6. The number of bacteria per ml is determined by measuring the OD600nm of 1 ml of the culture.
Note: The typical value for E. coli is 1 unit of the OD600nm corresponds to 5 x 108 bacteria/ml. - HeLa cells preparation
- Before infection, eukaryotic cells are cautiously washed once with pre-warmed (37 °C) HBSS.
- 100 μl of DMEM-HEPES medium is added in all the wells, except the 2 first rows, corresponding to the highest number of bacteria per cell (maximum multiplicity of infection or MOI) where 200 μl of DMEM-HEPES medium is added (Figure 1).
- Before infection, eukaryotic cells are cautiously washed once with pre-warmed (37 °C) HBSS.
- Infection assay
- After determination of the multiplicity of infection (MOI = number of bacteria per HeLa cell at the onset of the infection), the appropriate number of bacteria is added in the first 2 rows of the plate (containing 200 μl DMEM-HEPES medium).
- 2-fold serial dilution is performed to create an infectious dose-effect, in a final DMEM-HEPES medium volume of 100 μl (Figure 1).
Note: As an example, for an MOI of 50, add 50 x 1.5 x 104 = 75 x 104 bacteria for a well containing 1.5 x 104 HeLa cells. The number of dispensed cells is taken into account for the MOI calculation. - The plate is incubated for 4 h in a cell culture incubator.
- After determination of the multiplicity of infection (MOI = number of bacteria per HeLa cell at the onset of the infection), the appropriate number of bacteria is added in the first 2 rows of the plate (containing 200 μl DMEM-HEPES medium).
- End of the infection and post-infection incubation
- After infection of 4 h, the absorbance at 600 nm is measured with a microplate reader, to monitor bacterial growth. The typical values obtained are around 0.2-0.3 for MOI 50.
- Then, cells are washed carefully at least 3 times with an increasing volume of pre-warm (37 °C) HBSS (100 μl, 150 μl and finally 200 μl). The plate is observed under an inverted microscope to check whether most of the bacteria are removed. If required, additional washes can be performed.
- Finally, cells are incubated in 200 μl cell culture medium (see Recipes) for 3 h with 200 mg/ml gentamicin in a cell culture incubator.
- After infection of 4 h, the absorbance at 600 nm is measured with a microplate reader, to monitor bacterial growth. The typical values obtained are around 0.2-0.3 for MOI 50.
- Cells fixation
- The cells are washed carefully in cold PBS (100 μl) three times.
- 100 μl fixation solution (see Recipes) is added for 20 min under a chemical safety hood.
- The cells are washed carefully in cold PBS (100 μl) three times.
- Paraformaldehyde neutralization
- After the fixation step, cells are washed with PBS (100 μl per well), for 5 min under rapid agitation (variable speed shaker, 20-25 rpm). This washing step is repeated 3 times.
- Then, the paraformaldehyde is neutralized by adding 50 μl of neutralization solution (see Recipes) for 2 min under slow agitation (3-5 rpm).
- After the fixation step, cells are washed with PBS (100 μl per well), for 5 min under rapid agitation (variable speed shaker, 20-25 rpm). This washing step is repeated 3 times.
- Cells permeabilization
- The cells are washed 3 times for 5 min with PBS, 100 μl per well, under rapid agitation (20-25 rpm) as previously (Step 6).
- Then cells are permeabilized with cold permeabilization solution (see Recipes), 50 μl per well for 5 min, under slow agitation.
- The cells are washed 3 times for 5 min with PBS, 100 μl per well, under rapid agitation (20-25 rpm) as previously (Step 6).
- Blocking step
- Cells are washed 3 times with PST buffer (see Recipes), 100 μl per well, under agitation (20-25 rpm).
- Then, 50 μl/well of blocking solution (see Recipes) is added, and the plate is incubated at room temperature for 60 min under agitation (3-5 rpm).
- Cells are washed 3 times with PST buffer (see Recipes), 100 μl per well, under agitation (20-25 rpm).
- Anti-γ-H2AX immunostaining step
- Before immunostaining, cells are washed with PST buffer 3 times, 100 μl for 5 min under agitation (20-25 rpm).
- 25 μl per well of anti-γ-H2AX solution (see Recipes) is added and the plate is incubated for 2 h at room temperature or overnight at 4 °C (in that case, the plate has to be sealed carefully with Parafilm to avoid drying), under agitation (3-5 rpm).
Note: This solution is not added in the controls A and C (Figure 1), 25 μl of PST buffer is added instead during the incubation time.
- Before immunostaining, cells are washed with PST buffer 3 times, 100 μl for 5 min under agitation (20-25 rpm).
- Secondary detection step
- Cells are first washed 3 times with PST buffer, 100 μl per well for 5 min under agitation (20-25 rpm).
- Secondary detection is carried out using an infrared fluorescent dye conjugated IRDyeTM 800CW-conjugated goat anti-rabbit antibodies absorbing at 800 nm (1/1,000) in PST buffer. For DNA labeling, 1/1,000 dilution of RedDotTM2 in PST is used together with the secondary antibody (see secondary antibody solution in Recipes), 25 μl per well in every well of the plate.
- The plate is incubated in the dark for 1 h under agitation (3-5 rpm).
- Cells are first washed 3 times with PST buffer, 100 μl per well for 5 min under agitation (20-25 rpm).
- Plate scanning step
- The plate is washed 3 times in PST buffer, 100 μl per well for 5 min under agitation (20-25 rpm) in obscurity (covered with aluminum foil).
- Then, the plate is dried by reversing and patting it delicately on a tissue.
- The DNA and the γ-H2AX were simultaneously visualized using an Odyssey Infrared Imaging Scanner with the 680 nm fluorophore (red color) and the 800 nm fluorophore (green dye), respectively (Figure 3A).
- The plate is washed 3 times in PST buffer, 100 μl per well for 5 min under agitation (20-25 rpm) in obscurity (covered with aluminum foil).
Data analysis
A quantitative analysis of the frequency of DNA double-strand breaks can be performed using the fluorescence values (in RFU) obtained from the scans (Figures 2 and 3B). Firstly, the average γ-H2AX fluorescence per cell is obtained by dividing the measured γ-H2AX fluorescence per well (at 800 nm) by the corresponding measured fluorescence for RedDot-labelled DNA per well (at 680 nm) (Figure 2). The value for γ-H2AX fluorescence per cell is divided by the average fluorescence value at 800 nm obtained for the two wells serving as vehicle control (here corresponding to the wells marked ‘C’ in Figures 1 and 2) to determine the percent change in phosphorylation of H2AX levels relative to the vehicle control. The obtained number is defined as a genotoxic index (or γ-H2AX fold induction) and typically ranges from 0 to 2 in completely healthy cells, to 5 to 7 in cells infected with colibactin-producing E. coli at MOI 25 (Figure 2 and Figure 3B).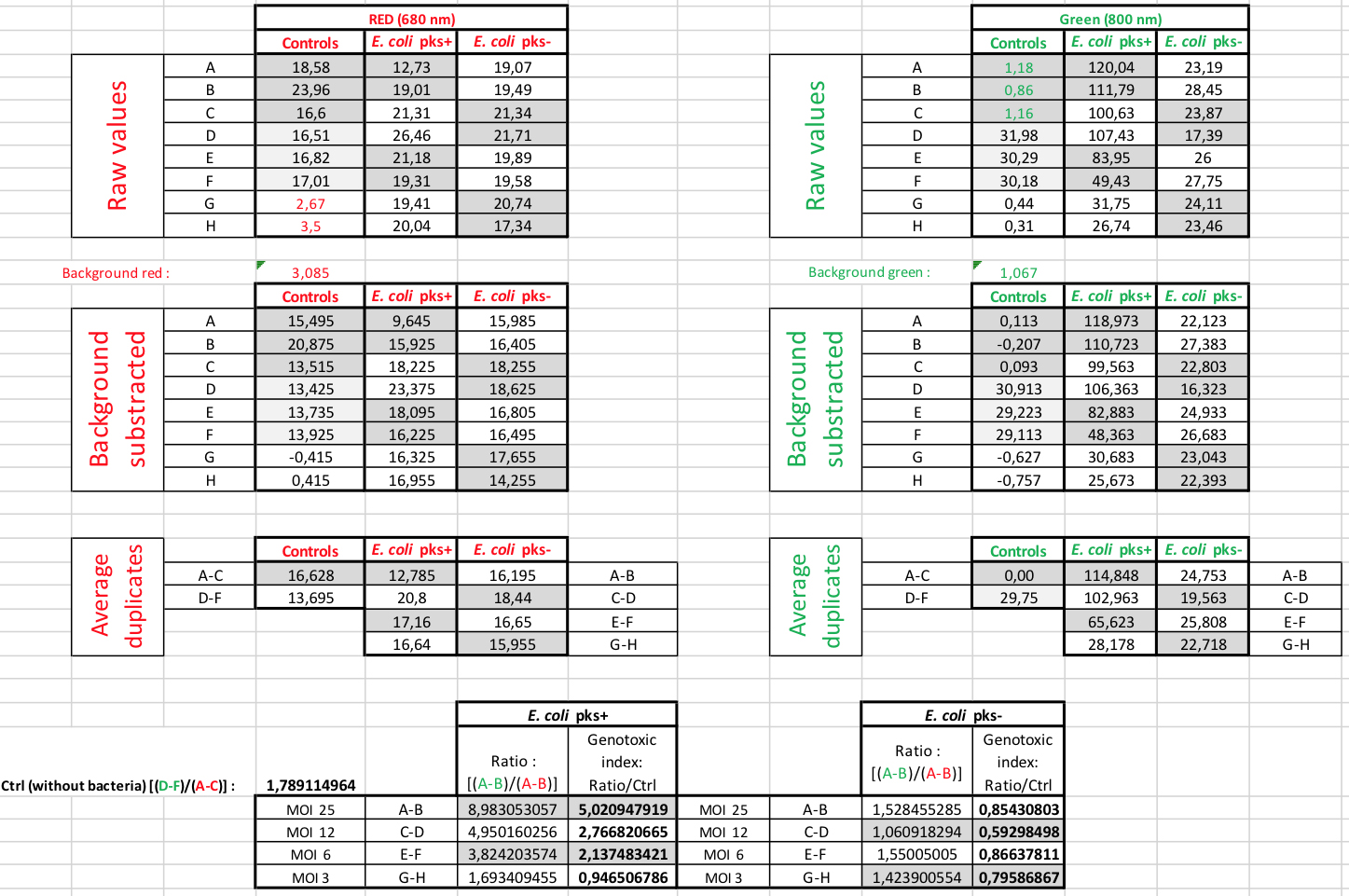
Figure 2. Data analysis of γ-H2AX quantification. Representative image of how we process data from the raw numbers to the calculation of the γ-H2AX fold induction (genotoxic index). 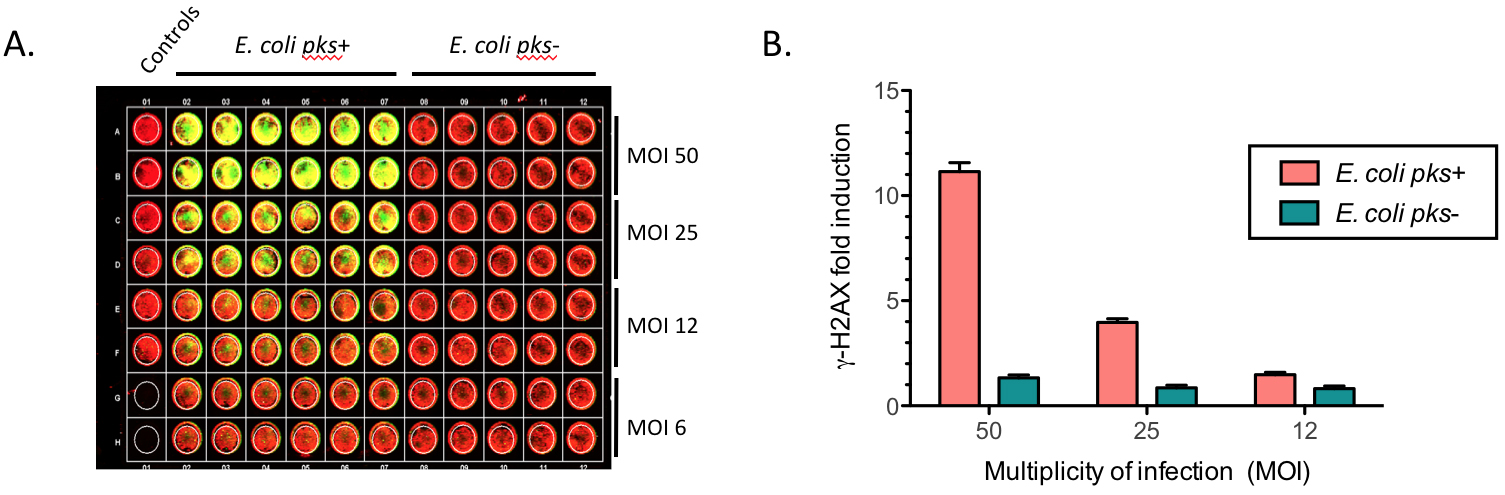
Figure 3. Quantification of phosphorylated H2AX (γ-H2AX) from infected HeLa cells with pks+ E. coli or with pks- E. coli at various MOI. A. Example of an In-Cell Western plate image with merged detection of total DNA (red, 680 nm) and γ-H2AX (green, 800 nm). A high level of γ-H2AX can be observed at high MOI for the cells infected with E. coli pks+. B. The γ-H2AX fold induction (γ-H2AX fluorescence normalized to the amount of cells per well), calculated relative to the control (non-infected cells), reveals a genotoxic dose-response depending on the MOI in cells infected by colibactin-producing bacteria (E. coli pks+) compared to cells infected by non colibactin-producing bacteria (E. coli pks-). Data represented in the graph were obtained from two biological replicates and two independent experiments. Data were plotted using GraphPad Prism 6.0. The mean with standard deviation (sd) is shown.
Recipes
- HeLa cells culture medium
450 ml DMEM high glucose with pyruvate and GlutaMax
50 ml fetal bovine serum
5 ml NEAA - Fixation solution (1x PBS, 4% PFA)
5 ml PFA stock solution
5 ml 20% 10x PBS
40 ml distilled water
Aliquot and store at -20 °C
Defreeze one aliquot the day of the experiment - Neutralization solution (20 mM NH4Cl in PBS)
a.Prepare the stock solution (1 M NH4Cl) in distilled water and store at 4 °C
b.Dissolve the stock solution in PBS to reach the appropriate concentration
c.Keep on ice until use - 10% Triton X-100
500 μl Triton X-100 in 50 ml PBS
This solution can be stored at 4 °C for 2-3 weeks - Permeabilization solution (1x PBS, 0.2% Triton X-100)
200 μl 10% Triton X-100
10 ml PBS
Keep on ice until use - Blocking solution (MAXblock blocking medium, 1x PHOSTOP inhibitor, RNase [100 μg/ml])
4.5 ml MAXblock blocking medium
500 μl 10x PHOSTOP inhibitor
50 μl RNase stock solution (10 mg/ml in PBS, 10 mM HEPES pH 7.5, 15 μM NaCl, aliquot stored at -20 °C)
Keep on ice until use - PST buffer (PBS, 2% fetal calf serum, 0.2% Triton X-100)
48 ml PBS
1 ml 2% fetal calf serum
1 ml 0.2% Triton X-100 in PBS - 100x azide
Dissolve sodium azide in distilled water at a final concentration of 50 mg/ml (5% w/w), store at 4 °C - Anti-γ-H2AX solution (rabbit monoclonal anti-γ-H2AX [1/200] in PST buffer, 500 μg/ml azide)
1.5 ml PST buffer
7.5 μl anti-γ-H2AX
15 μl 100x azide
Note: This solution can be reused 2-3 times, and stored at 5 °C for several weeks. - Secondary antibody solution (IRDyeTM 800CW Goat Anti-Rabbit [1/1,000], RedDotTM2 [1/1,000] in PST buffer)
2.5 ml PST
2.5 μl IRDyeTM 800CW Goat Anti-Rabbit
2.5 μl RedDotTM2
Acknowledgments
This protocol was adapted from previously published work (Audebert et al., 2010; Martin et al., 2013; Bossuet-Greif et al., 2016; Tronnet et al., 2017). This work was supported by the Agence Nationale de la Recherche (France) [grant ANR-13-BSV3–0015-02, ANR-13-BSV1–0028-01]. The authors declare no conflicts of interest or competing interests, regarding the publication of this protocol.
References
- Audebert, M. Riu, A., Jacques, C., Hillenweck, A., Jamin, E. L., Zalko, D., Cravedi, J. P. (2010). Use of the gammaH2AX assay for assessing the genotoxicity of polycyclic aromatic hydrocarbons in human cell lines. Toxicol Lett 199: 182-192.
- Bossuet-Greif, N., Belloy, M., Boury, M., Oswald, E. and Nougayrede, J. (2017). Protocol for HeLa cells infection with Escherichia coli strains producing colibactin and quantification of the induced DNA-damage. Bio-protocol 7(16): e2520.
- Bossuet-Greif, N., Dubois, D., Petit, C., Tronnet, S., Martin, P., Bonnet, R., Oswald, E. and Nougayrede, J. P. (2016). Escherichia coli ClbS is a colibactin resistance protein. Mol Microbiol 99(5): 897-908.
- Martin, P., Marcq, I., Magistro, G., Penary, M., Garcie, C., Payros, D., Boury, M., Olier, M., Nougayrede, J. P., Audebert, M., Chalut, C., Schubert, S. and Oswald, E. (2013). Interplay between siderophores and colibactin genotoxin biosynthetic pathways in Escherichia coli. PLoS Pathog 9(7): e1003437.
- Nougayrède, J. P., Homburg, S., Taieb, F., Boury, M., Brzuszkiewicz, E., Gottschalk, G., Buchrieser, C., Hacker, J., Dobrindt, U. and Oswald, E. (2006). Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313(5788): 848-851.
- Taieb, F., Petit, C., Nougayrede, J. P. and Oswald, E. (2016). The enterobacterial genotoxins: cytolethal distending toxin and colibactin. EcoSal Plus 7(1).
- Tronnet, S., Garcie, C., Brachmann, A. O., Piel, J., Oswald, E. and Martin, P. (2017). High iron supply inhibits the synthesis of the genotoxin colibactin by pathogenic Escherichia coli through a non-canonical Fur/RyhB-mediated pathway. Pathog Dis 75(5).
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Tronnet, S. and Oswald, E. (2018). Quantification of Colibactin-associated Genotoxicity in HeLa Cells by In Cell Western (ICW) Using γ-H2AX as a Marker. Bio-protocol 8(6): e2771. DOI: 10.21769/BioProtoc.2771.
Category
Microbiology > Microbe-host interactions > In vitro model > Cell line
Cell Biology > Cell staining > Protein
Biochemistry > Protein > Fluorescence
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










