- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
RNA Cap Methyltransferase Activity Assay
Published: Vol 8, Iss 6, Mar 20, 2018 DOI: 10.21769/BioProtoc.2767 Views: 8453
Reviewed by: Gal HaimovichAmanda GarnerC. Kiong Ho

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In vitro Assays for the Detection of Calreticulin Exposure, ATP and HMGB1 Release upon Cell Death
Yuting Ma and Heng Yang
Dec 20, 2016 19994 Views

Analyzing (Re)Capping of mRNA Using Transcript Specific 5' End Sequencing
Daniel del Valle Morales and Daniel R. Schoenberg
Oct 20, 2020 4306 Views

Measuring in vitro ATPase Activity with High Sensitivity Using Radiolabeled ATP
Sarina Veit and Thomas Günther Pomorski
May 20, 2023 2273 Views
Abstract
Methyltransferases that methylate the guanine-N7 position of the mRNA 5’ cap structure are ubiquitous among eukaryotes and commonly encoded by viruses. Here we provide a detailed protocol for the biochemical analysis of RNA cap methyltransferase activity of biological samples. This assay involves incubation of cap-methyltransferase-containing samples with a [32P]G-capped RNA substrate and S-adenosylmethionine (SAM) to produce RNAs with N7-methylated caps. The extent of cap methylation is then determined by P1 nuclease digestion, thin-layer chromatography (TLC), and phosphorimaging. The protocol described here includes additional steps for generating the [32P]G-capped RNA substrate and for preparing nuclear and cytoplasmic extracts from mammalian cells. This assay is also applicable to analyzing the cap methyltransferase activity of other biological samples, including recombinant protein preparations and fractions from analytical separations and immunoprecipitation/pulldown experiments.
Keywords: RNABackground
The N7-methylguanosine cap at the 5’ end of an mRNA is a modification essential for proper eukaryotic mRNA processing, localization, and translation. The N7 methyl group is particularly critical for the mRNA life cycle, as it drastically increases the binding affinity of cap-binding proteins (Niedzwiecka et al., 2002) and protects mRNAs from cap-quality control surveillance mechanisms (Jiao et al., 2013). We recently reported that the mammalian RNA guanine-7 methyltransferase (RNMT) functions beyond its canonical role in nuclear co-transcriptional cap synthesis to participate in cytoplasmic RNA recapping (Trotman et al., 2017). We used the protocol presented here to demonstrate that the cap methyltransferase activity of cytoplasmic RNMT is unexpectedly robust relative to nuclear RNMT. Additionally, siRNA-mediated knockdown of RNMT greatly reduced the cap methyltransferase activity of cytoplasmic extracts, suggesting that RNMT is the predominant, if not only cap methyltransferase in the cytoplasm of mammalian cells. Nuclear RNMT exists as a heterodimer with RNMT-activating miniprotein (RAM, Gonatopoulos-Pournatzis et al., 2011), and we demonstrated that cytoplasmic RNMT also binds to RAM. Reduced cytoplasmic cap methyltransferase activity upon RAM knockdown indicated that RAM is a required cofactor for cytoplasmic RNMT.
This protocol is adapted from two earlier publications characterizing human RNMT (Cowling, 2010; Pillutla et al., 1998), with modifications that standardize generation of the substrate RNA, avoid cumbersome phenol-chloroform extractions with radioactive samples, and enable quantification of cap methyltransferase activity. We note that an alternative, nonradioactive assay has been reported for the analysis of cap methyltransferase reactions (Peyrane et al., 2007), but this method requires HPLC instrumentation that may not be available to all labs and may differ from the one presently reported in terms of sensitivity and sample compatibility. Additionally, a fluorescent assay for measuring cap methyltransferase activity was recently described (Aouadi et al., 2017), but this assay indirectly measures activity by monitoring the accumulation of S-adenosylhomocysteine (SAH) and may be incompatible with biological samples containing SAH. We hope that the level of detail in the protocol presented here enables future investigators to easily repeat and build upon our work.
Materials and Reagents
- 0.2 ml PCR tubes (e.g., BioExpress, GeneMate, catalog number: T-3225-1 )
- NucAway Spin Columns (Thermo Fisher Scientific, InvitrogenTM, catalog number: AM10070 )
- 1.7 ml plastic, sterile, RNase-free microcentrifuge tubes (e.g., BioExpress, GeneMate, catalog number: C-3262-1 )
- 10 cm or 15 cm culture dishes (e.g., Alkali Scientific, catalog numbers: TD0100 , TD0150 )
- Sterile nitrile gloves
- Cell lifters (e.g., BioExpress, GeneMate, catalog number: T-2443-4 )
- Sterile, RNase-free tips for P10, P20, P100, and P1000 pipets
- Plastic, disposable cuvettes for Bradford assay (e.g., Fisher Scientific, catalog number: 14-955-127 )
- Plastic wrap (e.g., Saran or Stretch-Tite brand)
- Paper labeling tape (e.g., Fisher Scientific, catalog number: 15-901-20H)
Manufacturer: Nevs, catalog number: 1590120H . - Polyethylenimine (PEI) cellulose TLC plates (Macherey-Nagel, catalog number: 801053 ; see Note 2)
- Immobilon-FL polyvinylidene difluoride (PVDF) membrane (Merck, catalog number: IPFL00010 )
- Scintillation vials compatible with liquid scintillation counter (e.g., DWK Life Sciences, WHEATON, catalog number: 225414 )
- U2OS cells (ATCC, catalog number: HTB-96 ) or HEK293 cells (ATCC, catalog number: CRL-1573 )
- P1 nuclease (United States Biological, catalog number: N7000 ), resuspended in RNase-free water to 0.625 U/μl
- Cap analog GpppG (New England Biolabs, catalog number: S1407 ), resuspended in RNase-free water to 10 mM
- Cap analog m7GpppG (New England Biolabs, catalog number: S1404 ), resuspended in RNase-free water to 10 mM
- RNase-free water (e.g., from a Millipore Synergy water purification system, 18.2 MΩ cm)
- Single-stranded DNA sense oligo
- Single-stranded DNA antisense oligo
- 5’ CATGCAAATTAACCCTCACTAAAGGGAGACCGGAATTCGAGCTCGCCCGGGGATC 3’ for T3 transcription template, resuspended in RNase-free water to 100 μM (e.g., synthesized by Integrated DNA Technologies (IDT); underlined is a T3 promoter sequence, bold sequence matches the transcribed 32-nucleotide (nt) pppRNA)
- 5’ GATCCCCGGGCGAGCTCGAATTCCGGTCTCCCTTTAGTGAGGGTTAATTTGCATG 3’ for T3 transcription template, resuspended in RNase-free water to 100 μM (e.g., synthesized by IDT)
- MEGAscript T3 Transcription Kit (Thermo Fisher Scientific, Ambion, catalog number: AM1138 )
- Pre-cast polyacrylamide mini-gels for urea polyacrylamide gel electrophoresis (urea-PAGE; e.g., Bio-Rad Laboratories, catalog number: 4566053 )
- Pre-cast polyacrylamide mini-gels for sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE; e.g., Bio-Rad Laboratories, catalog number: 4568094 )
- 2x Laemmli Sample Buffer (Bio-Rad Laboratories, catalog number: 1610737 )
- SYBR Gold Nucleic Acid Gel Stain (Thermo Fisher Scientific, InvitrogenTM, catalog number: S11494 )
- RNA size marker (e.g., Thermo Fisher Scientific, InvitrogenTM, catalog number: AM7778 )
- 2x RNA loading dye (e.g., Thermo Fisher Scientific, Thermo ScientificTM, catalog number: R0641 )
- 40 U/μl RNaseOUT Recombinant Ribonuclease Inhibitor (Thermo Fisher Scientific, InvitrogenTM, catalog number: 10777019 )
- Recombinant triphosphatase-guanylyltransferase capping enzyme (see Note 1)
- [α-32P]GTP (3,000 Ci/mmol, PerkinElmer, catalog number: BLU506H250UC )
- RNA Clean & Concentrator-5 kit (Zymo Research, catalog number: R1016 )
- ScintiSafe Econo 1 scintillation fluid (Fisher Scientific, catalog number: SX20-5 )
Note: This product has been discontinued. - McCoy’s 5A medium (for U2OS cells; Thermo Fisher Scientific, GibcoTM, catalog number: 16600082 )
- Dulbecco’s modified Eagle medium (DMEM; for HEK293 cells; Thermo Fisher Scientific, GibcoTM, catalog number: 21013024 )
- Fetal bovine serum (FBS; Atlanta Biologicals, catalog number: S10350 )
- Phosphate-buffered saline (PBS; e.g., Thermo Fisher Scientific, GibcoTM, catalog number: 10010049 )
- Bio-Rad Protein Assay Dye Reagent Concentrate (Bio-Rad Laboratories, catalog number: 5000006 )
- Pre-stained protein marker (e.g., Bio-Rad Laboratories, catalog number: 1610393 )
- Bovine serum albumin (BSA; e.g., Fisher Scientific, catalog number: BP1600-100 ) dissolved in water to 1 μg/μl
- Antibody toward nuclear protein for Western blotting (e.g., rabbit polyclonal anti-nucleolin antibody, 1:5,000 working dilution, Sigma-Aldrich, catalog number: N2662 )
- Antibody toward cytoplasmic protein for Western blotting (e.g., mouse monoclonal anti-α-tubulin, 1:10,000 working dilution, Sigma-Aldrich, catalog number: T6199 )
- Appropriate secondary antibodies (e.g., for infrared imaging, Thermo Fisher Scientific, Invitrogen, catalog numbers: A-21109 and A-21058 , at 1:10,000 working dilutions)
- (Optional) Vaccinia Capping Enzyme (New England Biolabs, catalog number: M2080S )
- Dithiothreitol (DTT, Thermo Fisher Scientific, Thermo ScientificTM, catalog number: R0861 )
- RNase-free 10x Tris/borate/EDTA (TBE) buffer (e.g., Thermo Fisher Scientific, InvitrogenTM, catalog number: 15581 )
- Phenylmethylsulfonyl fluoride (PMSF; Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 36978 )
- Isopropanol
- Tris (e.g., VWR, AMRESCO, catalog number: 0497 )
- Glycine (e.g., VWR, AMRESCO, catalog number: 0167 )
- SDS
- Methanol (e.g., Fisher Scientific, catalog number: A452SK-4 )
- Sodium chloride (NaCl; e.g., Fisher Scientific, catalog number: BP358 )
- 5 M NaCl (e.g., Thermo Fisher Scientific, catalog number: AM9759 )
- Concentrated HCl
- Tween 20 (e.g., Fisher Scientific, catalog number: BP337 )
- 32 mM S-adenosylmethionine (SAM; New England Biolabs, catalog number: B9003 )
- 3 M sodium acetate, pH 5.2 (e.g., Thermo Fisher Scientific, Thermo ScientificTM, catalog number: R1181 )
- Ammonium sulfate (e.g., Sigma-Aldrich, catalog number: A4418 )
- IGEPAL CA-630 (Sigma-Aldrich, catalog number: I8896 )
- 1 M Tris-HCl, pH 7.5 (e.g., AMRESCO, catalog number: E691 )
- 1 M magnesium chloride (MgCl2, e.g., Thermo Fisher Scientific, InvitrogenTM, catalog number: AM9530G )
- 500 mM ethylenediaminetetraacetic acid, pH 8.0 (EDTA; e.g., Thermo Fisher Scientific, InvitrogenTM, catalog number: 15575020 )
- Glycerol (e.g., Fisher Scientific, catalog number: BP229-1 )
- 1 M HEPES pH 7.3 (e.g., AMRESCO, catalog number: J848 )
- 2 M potassium chloride (KCl; e.g., Alfa Aesar, catalog number: J75896 )
- Protease inhibitor cocktail (Sigma-Aldrich, catalog number: P8340 )
- Phosphatase inhibitor cocktail 2 (Sigma-Aldrich, catalog number: P5726 )
- Phosphatase inhibitor cocktail 3 (Sigma-Aldrich, catalog number: P0044 )
- 1 M DTT (see Recipes)
- 1x Tris-buffered saline (TBS; see Recipes)
- 1x TBE (running buffer for urea-PAGE) (see Recipes)
- 20 mM DTT (see Recipes)
- 100 mM PMSF in isopropanol (see Recipes)
- 10x Tris/glycine (see Recipes)
- 10% (w/v) SDS (see Recipes)
- Tris/glycine/SDS running buffer for SDS-PAGE (see Recipes)
- Tris/glycine/methanol/SDS transfer buffer (see Recipes)
- 20x Tris-buffered saline (TBS) (see Recipes)
- 3% BSA in TBS (see Recipes)
- 40% (v/v) Tween 20 (see Recipes)
- TBS-T (see Recipes)
- 1 μM SAM (see Recipes)
- 500 mM sodium acetate, pH 5.2 (see Recipes)
- 0.4 M ammonium sulfate
- 10% IGEPAL CA-630 (v/v) in water (see Recipes)
- 10x annealing buffer (see Recipes)
- 4x capping buffer (see Recipes)
- YO Lysis Buffer (see Recipes)
- YO Buffer A (see Recipes)
- 10x cap methylation buffer (see Recipes)
Equipment
- Eye protection
- NanoDrop spectrophotometer (Thermo Fisher, model: NanoDropTM 1000 , catalog number: ND-1000)
- Lucite/Plexiglass acrylic benchtop shielding for handling 32P (e.g., Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 6700-2418 )
- Geiger counter
- Metric (centimeter) ruler
- Scissors (e.g., Westcott, catalog number: ACM44217 )
- Graphite pencil
- Long (at least 18 cm) forceps or tongs (e.g., Fisher Scientific, catalog number: 15-186 )
- P10, P20, P100, and P1000 pipets
- Thermal cycler (e.g., MJ Research, model: PTC-200 )
- Heating block (e.g., Bioer, model: MB 101 )
- -80 °C freezer
- Handheld 254 nm UV light (e.g., UVP, model: 95-0016-14 )
- Liquid scintillation counter (e.g., Beckman Coulter, model: LS 6000IC )
- Water-jacketed incubator (e.g., Thermo Fisher Scientific, Thermo ScientificTM, model: Forma® Series II) set to 37 °C with 5% CO2
- Refrigerated centrifuge (Eppendorf, model: 5415 R )
- Programmable rotator-mixer (Grant Instruments, model: PTR-30 ) at 4 °C and set to 10 rpm orbital rotation
- UV-vis spectrophotometer (e.g., Beckman Coulter, model: DU 640 ) set to 595 nm
- Electrophoresis system for PAGE and membrane transfer (e.g., Bio-Rad Laboratories, catalog number: 1660828EDU )
- Opaque western blot incubation boxes (e.g., LI-COR, catalog number: 929-97205 )
- Western blot imaging system (e.g., LI-COR, model: Odyssey Imaging Systems )
- Rectangular glass TLC chamber (e.g., Miles Scientific, model: A70-22 , formerly Analtech)
- Electric hair dryer (optional)
- Storage phosphor screen (e.g., GE/Amersham Biosciences)
- Light eraser for storage phosphor screen (e.g., Molecular Dynamics Image Eraser)
- Typhoon imaging system (e.g., GE Healthcare, Amersham Biosciences, model: Typhoon 9200 )
Software
- Microsoft Excel software
- ImageQuant TL software
Procedure
A schematic overview of this protocol is presented in Figure 1. In short, an in-vitro-transcribed short RNA is guanylylated using [α-32P]GTP to generate a Gp*ppRNA substrate, with the radiolabeled phosphate indicated by an asterisk. This GpppRNA substrate is briefly incubated with nuclear or cytoplasmic extracts (or other biological samples) and SAM. Different levels of cap methyltransferase activity in each extract will result in differential N7-methylation of the GpppRNA substrate to produce m7GpppRNA. The mixtures of GpppRNA and m7GpppRNA are then digested to individual nucleotides with P1 nuclease, leaving intact the radiolabeled cap dinucleotide structures that are finally analyzed by TLC and phosphorimaging.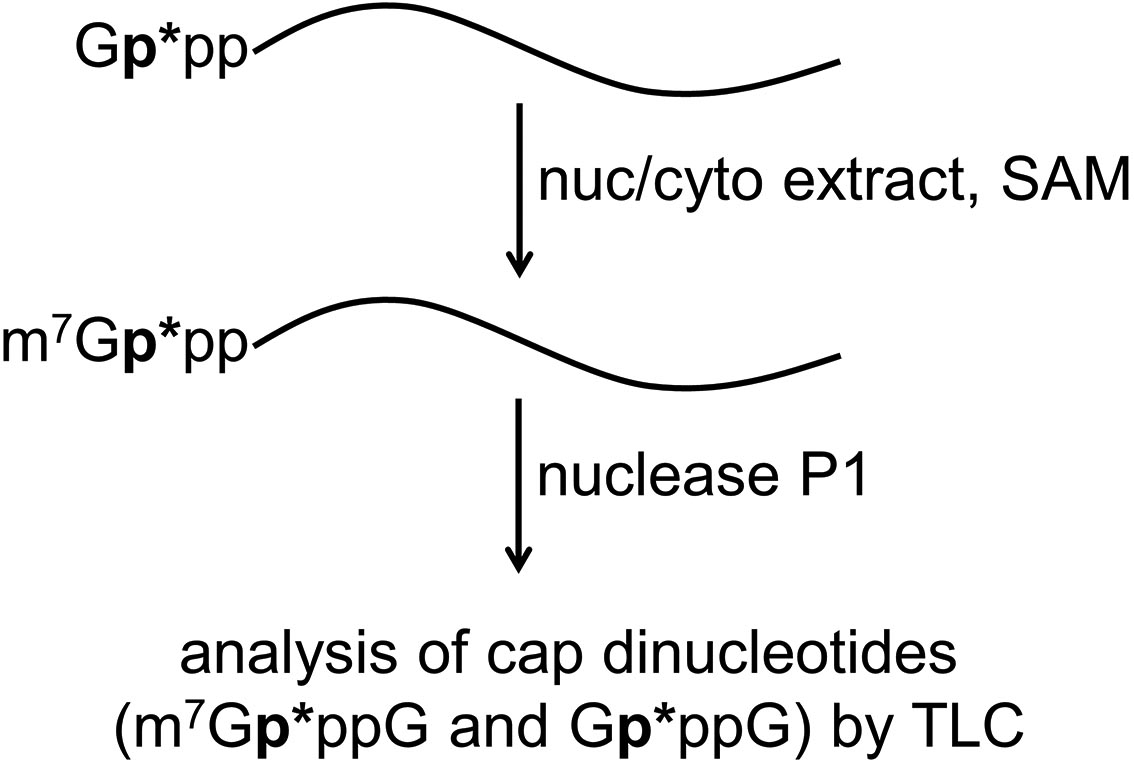
Figure 1. Schematic overview of cap methyltransferase activity assay
- Preparation of radiolabeled GpppRNA substrate for cap methyltransferase activity assay (see Note 3)
- Prepare a 30 μl mixture in a 0.2 ml PCR tube by combining the following: 15 μl of RNase-free water, 3 μl of 10x annealing buffer (see Recipes), 6 μl of 100 μM ssDNA sense oligo, 6 μl of 100 μM ssDNA antisense oligo.
- To denature and then slowly anneal the two oligos to create the dsDNA template for T3 transcription, place the mixture on a thermal cycler at 98 °C for 2 min, and ramp the temperature from 98 °C to 20 °C in 60 min (1.3 °C per min, down to 20 °C). Transfer this tube of dsDNA onto ice, and in a separate tube, prepare a 1 μM (1 pmol/μl) working stock by diluting it 20-fold with RNase-free water.
- Using the components provided in the MEGAscript T3 Transcription Kit, prepare a 20 μl reaction in a 0.2 ml PCR tube by combining in the following order: 6 μl of RNase-free water, 2 μl of 75 mM ATP, 2 μl of 75 mM CTP, 2 μl of 75 mM GTP, 2 μl of 75 mM UTP, 2 μl of 10x T3 reaction buffer, 2 μl of 1 μM dsDNA template, 2 μl of T3 RNA polymerase enzyme mix. Gently mix, and incubate at 37 °C for 16-24 h. This prolonged reaction time enables transcription of high yields of 32 nt 5’ triphosphate RNA (pppRNA).
- Bring the T3 transcription reaction product mixture to 50 μl by adding 30 μl of RNase-free water, and purify using a NucAway Spin Column according to manufacturer’s instructions.
- Measure the concentration of the purified pppRNA using a NanoDrop. In our experience, at least 7-8 μg of pppRNA should be recovered after a 24 h transcription reaction. Calculate the molar concentration of the pppRNA assuming a molecular mass of 10,830.3 g/mol.
- Further assess the purity of the pppRNA by 15% polyacrylamide urea-PAGE. As little as 150 fmol (1.6 ng) of pppRNA per lane should be sufficient to visualize under UV light after post-staining the gel with SYBR Gold at a dilution of 1:20,000 in 1x TBE. The pppRNA should run as a single band at the expected size.
- The pppRNA should be stored in small aliquots at -20 °C or -80 °C to avoid multiple freeze-thaw cycles.
- To prepare a 40 μl capping (guanylylation) reaction, combine in order the following components in a 1.7 ml tube on ice: RNase-free water (for 40 μl total volume), 10 μl of 4x capping buffer (see Recipes), 1 μl of RNaseOUT, 4.8 pmol of 32 nt pppRNA, 2 pmol of recombinant capping enzyme (see Note 1), 24 pmol (total GTP) of [α-32P]GTP (see Note 4). Gently mix and incubate at 30 °C for 3 h, then heat-inactivate by incubating at 65 °C for 10 min.
- Transfer 1.33 μl (1/30) of the capping reaction product to a new tube on ice to save as ‘input’ for scintillation counting. Bring the capping reaction to 50 μl by adding 11.33 μl of RNase-free water. Purify the RNA using the Zymo RNA Clean & Concentrator-5 kit according to manufacturer’s instructions, using 30 μl of RNase-free water for the final elution step. Return the purified RNA to ice, and transfer 1 μl (1/30) to a new tube on ice for scintillation counting.
- Mix the set-aside 1/30 capping reaction ‘input’ and purified RNA samples each with 1.5 ml of ScintiSafe Econo 1 scintillation fluid and transfer to scintillation vials. Measure the radioactivity in each mixture by scintillation counting. Determine the concentration of purified [32P]G-capped RNA according to the following equation: [GpppRNA] (in μM) = (24 pmol) x (purified counts/input counts)/(30 μl).
- The purified [32P]G-capped RNA may be stored at -20 °C but should be used within a few days to limit radioactive decay. Be sure to account for radioactive decay when determining the appropriate amount to use in the cap methyltransferase activity assay.
- Prepare a 30 μl mixture in a 0.2 ml PCR tube by combining the following: 15 μl of RNase-free water, 3 μl of 10x annealing buffer (see Recipes), 6 μl of 100 μM ssDNA sense oligo, 6 μl of 100 μM ssDNA antisense oligo.
- Preparation of nuclear and cytoplasmic extracts of mammalian cells
- Two to three days in advance, seed 10-cm or 15-cm dishes with U2OS, HEK293, or other type of adherent mammalian cells such that the dishes will be ~80-90% confluent at the time of harvesting. U2OS cells should be grown in McCoy’s 5A medium containing 10% (v/v) FBS; HEK293 cells should be grown in DMEM containing 10% (v/v) FBS. Cells should be grown in an incubator set to 37 °C with 5% CO2.
- On the day of harvesting the cells, prepare YO Lysis Buffer and YO Buffer A (see Recipes) on ice, leaving out the DTT, PMSF, protease inhibitor cocktail, and phosphatase inhibitor cocktails until immediately before using the buffers.
- Remove culture medium from adherent cells, rinse with PBS and remove, and then harvest by scraping into 1 ml of PBS and transferring to 1.7 ml centrifuge tubes. Gently pellet the cells (70 x g for 10 min at 4 °C), and then carefully remove the supernatant PBS by aspiration. Make note of the approximate volume of the cell pellets, and place the tubes on ice.
- For the preparation of cytoplasmic extracts, immediately add YO Lysis Buffer equivalent to 4-5 times the volume of each cell pellet. Gently resuspend the cells with 10 up-down strokes of a P1000 pipet. Incubate the cells on ice for 10 min, and then mix with 5 additional up-down strokes of the P1000 pipet. Pellet nuclei (16,100 x g for 10 min at 4 °C), and then transfer the cytoplasmic extract supernatants to new, pre-chilled 1.7 ml tubes on ice.
- Carefully remove any residual cytoplasmic extract from the nuclear pellets using a P10 pipet. To prepare nuclear extracts, add 4-5 original-cell-pellet volumes of YO Buffer A, and resuspend each nuclear pellet with 15 up-down strokes of a P200 pipet. Incubate the tubes end-over-end at 4 °C for 20 min, and then pellet nuclear debris (16,100 x g for 5 min at 4 °C). Transfer the nuclear extract supernatants to new, pre-chilled 1.7 ml tubes on ice.
- Measure the concentration of protein in each extract by Bradford assay, using the Bio-Rad Protein Assay Dye Reagent Concentrate according to manufacturer’s instructions and using a standard curve of bovine serum albumin (BSA) at 0, 1, 2, 4, 6, and 8 μg/ml. Diluting the extracts 250- to 500-fold in the 1x dye reagent typically gives measurements within the linear range of the assay. Using a program such as Microsoft Excel, calculate the protein concentrations using the best-fit line of the BSA standard curve.
- For the cap methyltransferase activity assay, prepare 15-20 μl samples of each extract in new, pre-chilled tubes on ice. If comparing nuclear and cytoplasmic extracts, it is critical to ensure that these are in the same buffer composition. This can be accomplished by first bringing the extracts all to the same protein concentration (typically 0.5-1.0 μg/μl; such calculations can be greatly aided by software such as Microsoft Excel) with the same buffer used prior (i.e., YO Lysis Buffer for cytoplasmic extracts and YO Buffer A for nuclear extracts) and then mixing each sample with an equal volume of the other buffer (i.e., YO Buffer A for cytoplasmic extracts and YO Lysis Buffer for nuclear extracts). Store these extract samples in small aliquots at -80 °C.
- Assess the quality of subcellular fractionation by running the nuclear and cytoplasmic extracts on SDS-PAGE followed by Western blot analysis. The use of nuclear and cytoplasmic control antibodies (e.g., against nucleolin and tubulin) can determine whether cross-contamination occurred during the fractionation procedure. For an example of expected Western blot results, see Figure 1C of Trotman et al., 2017.
- Two to three days in advance, seed 10-cm or 15-cm dishes with U2OS, HEK293, or other type of adherent mammalian cells such that the dishes will be ~80-90% confluent at the time of harvesting. U2OS cells should be grown in McCoy’s 5A medium containing 10% (v/v) FBS; HEK293 cells should be grown in DMEM containing 10% (v/v) FBS. Cells should be grown in an incubator set to 37 °C with 5% CO2.
- Preparation of TLC plate for cap methyltransferase activity assay
- Wearing clean gloves, use a clean ruler and scissors to cut a 20 x 20 cm PEI cellulose TLC plate into four 10 x 10 cm squares. We recommend using clean forceps to handle the plate to limit contact with any contaminants that may be present on gloves. Store any unused plates at 4 °C. See Note 2 regarding TLC performance.
- Use the ruler and a pencil to lightly draw a faint, straight line (origin) 1.5 cm from the bottom edge of a 10 x 10 cm plate. Draw faint tick marks on the origin 1 cm apart to indicate where samples will be spotted. An example TLC plate is shown in Figure 2.
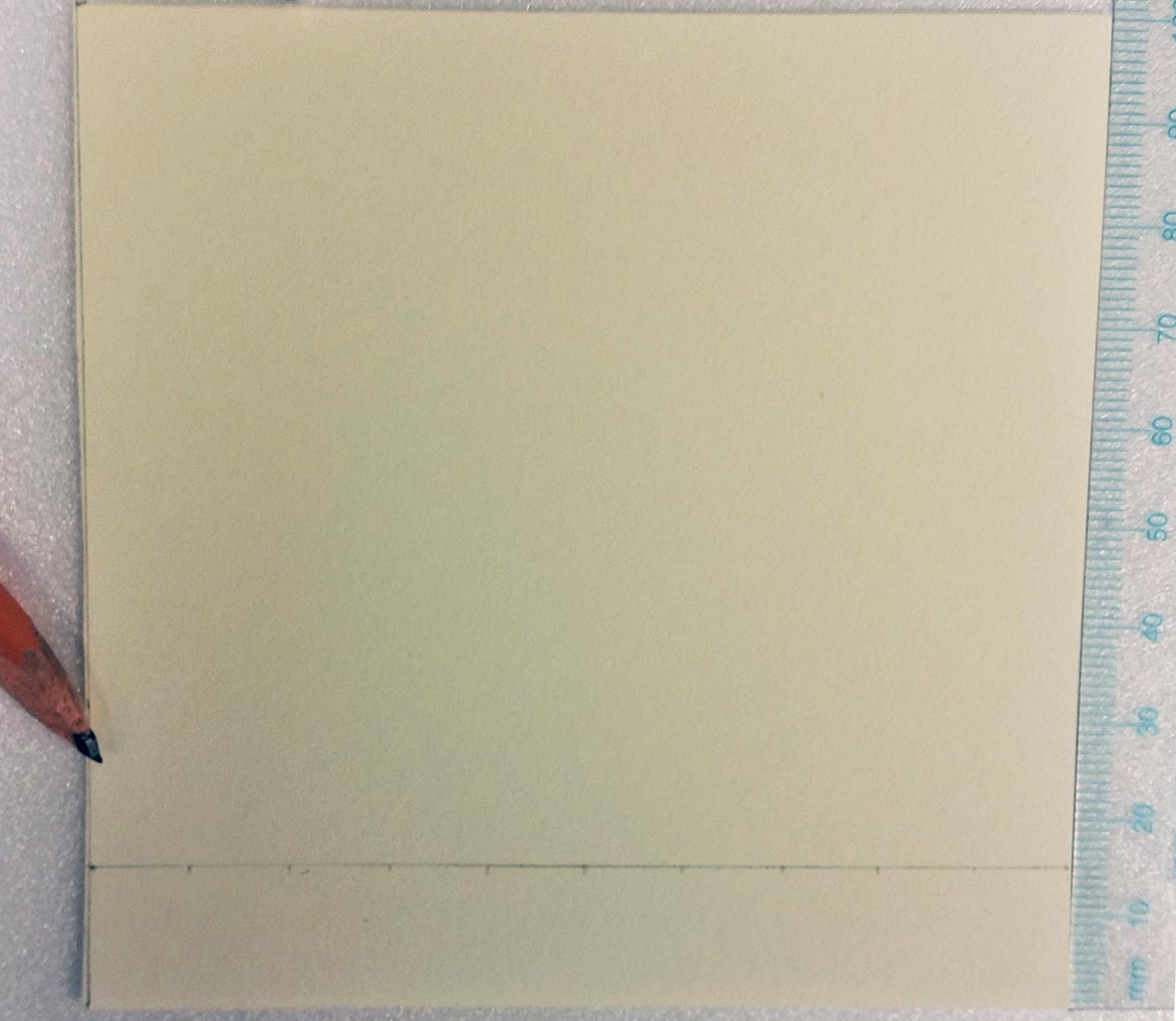
Figure 2. Example of TLC plate preparation - Pre-run the TLC plate with RNase-free water. To do so, fill a rectangular glass TLC chamber with just enough water to cover the bottom (approximately 60 ml in the model listed above) such that the water depth does not exceed 0.75 cm. Using long forceps, gently place the TLC plate upright in the water. Take care to ensure that the left and right edges enter the water simultaneously, as this will promote even migration of water up the TLC plate. This pre-running step forces contaminants to the top of the TLC plate and greatly improves performance for the subsequent chromatographic analysis of the cap methyltransferase activity assay samples.
- Once water has reached the top of the TLC plate, remove the TLC plate from the chamber and allow it to air-dry. Shaking the plate by hand or using a commercial hair dryer (without heating) can help speed up this process.
- Pre-run TLC plates can be stored at 4 °C for at least 2 weeks.
- Wearing clean gloves, use a clean ruler and scissors to cut a 20 x 20 cm PEI cellulose TLC plate into four 10 x 10 cm squares. We recommend using clean forceps to handle the plate to limit contact with any contaminants that may be present on gloves. Store any unused plates at 4 °C. See Note 2 regarding TLC performance.
- Cap methyltransferase activity assay
- After thawing components on ice, prepare 10 μl cap methyltransferase reactions in 1.7 ml tubes on ice by combining the following in order: RNase-free water (for 10 μl total volume), 1 μl of 10x cap methylation buffer (see Recipes), 1 μl of 1 μM SAM, 1 μl of 20 mM DTT, 1 μl of RNaseOUT, nuclear or cytoplasmic extract for 1 μg of total protein, 5 fmol of [32P]G-capped RNA. Using a master mix of the reaction components (omitting the extracts) is recommended if working with a large number of extract samples. Gently mix, incubate at 37 °C for 30 min, and then immediately return to ice.
- Add 40 μl of RNase-free water to each tube and purify the RNA samples using the Zymo Clean & Concentrator-5 kit according to manufacturer’s instructions, using 7 μl RNase-free water for the final elution step. Place the purified RNA on ice.
- Prepare 7 μl P1 nuclease digestion reactions in 1.7 ml tubes on ice by combining the following in order: 5.0 μl of purified RNA from cap methyltransferase reactions, 0.6 μl of RNase-free water, 0.7 μl of 500 mM sodium acetate pH 5.2, 0.7 μl of 0.625 U/μl P1 nuclease. Gently mix and incubate at 37 °C for 30 min. The samples may now be stored at -20 °C or analyzed immediately.
- On the 1 cm tick marks on the origin of a pre-run PEI cellulose TLC plate, carefully spot 1 μl of the P1 nuclease digestion products using a P10 pipet. Allow the spots to completely air-dry, and then spot an additional 1 μl of the P1 digestion products on the same positions as before. As controls, also spot 2 μl (1 μl at a time) of 10 mM GpppG and 10 mM m7GpppG standards on a separate tick mark or marks (see Note 5).
- After allowing the spots to completely air-dry, use long forceps to evenly place the TLC plate upright in a rectangular TLC chamber containing approximately 60 ml 0.4 M ammonium sulfate mobile phase. Allow the mobile phase to develop upward, approximately 6-7 cm beyond the origin, and then remove the TLC plate from the chamber. Allow the TLC plate to completely air-dry, which can be sped up by hand-shaking or using a hair dryer without heat.
- Wearing proper eye and skin protection, observe the TLC plate under a handheld 254 nm UV light to determine the positions of the GpppG and m7GpppG standards. These can be noted by marking their positions with a pencil or by taking a photo with a handheld camera.
- Wrap the TLC plate in plastic wrap so that the front side is covered by one layer. Place the TLC plate in a storage phosphor screen cassette, taping the top edge to the cassette to keep it in place. Place a recently light-erased storage phosphor screen over the TLC plate, close the cassette, and expose at room temp for at least 16 h for maximum signal.
- Remove the TLC plate from the cassette and image the storage phosphor screen with a Typhoon imager set to phosphor mode. Typical results are shown in Figure 3.
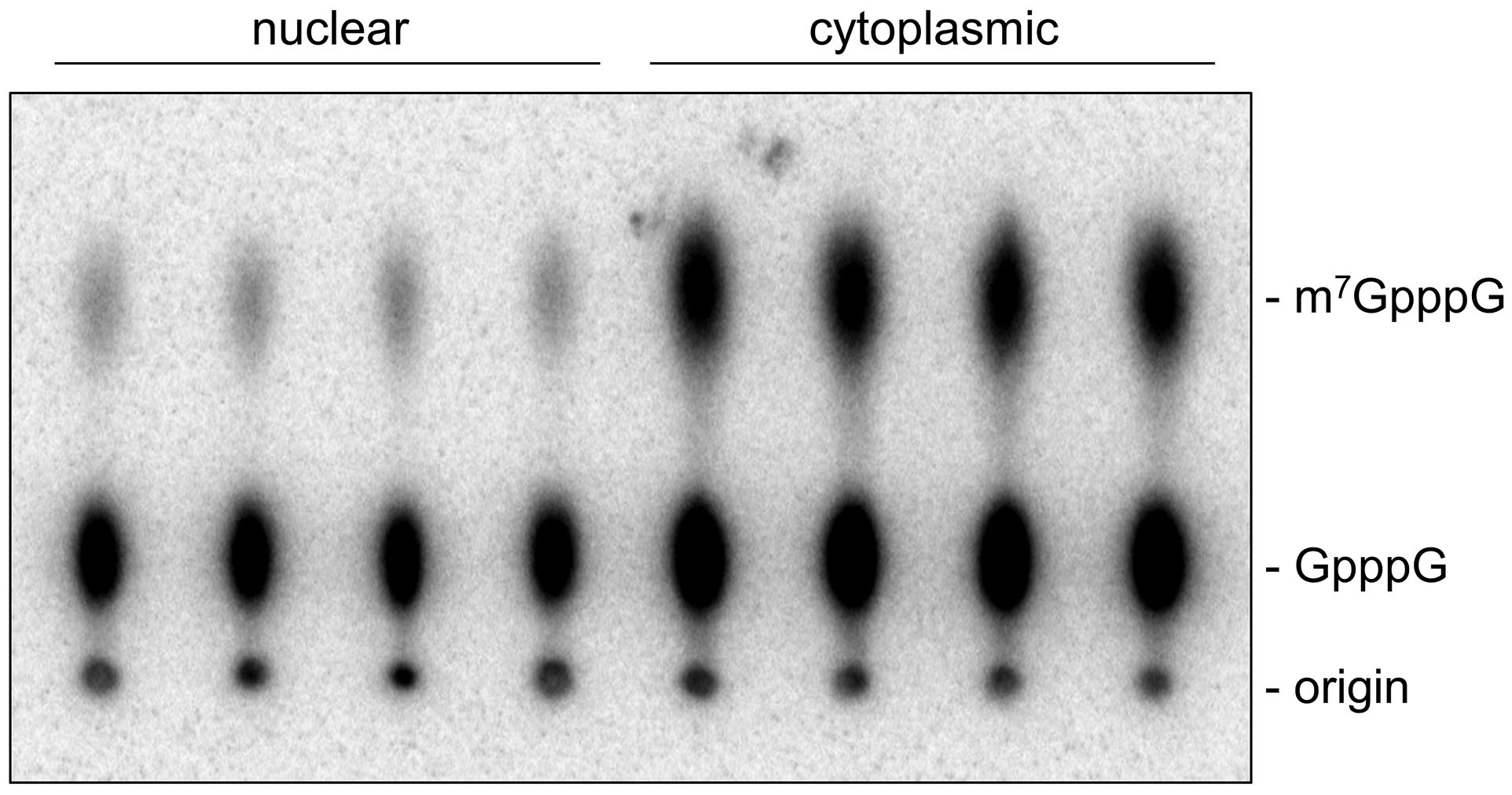
Figure 3. Typical phosphor image results. To demonstrate reproducibility, four technical replicates of the described activity assay were performed with U2OS cell nuclear and cytoplasmic extracts.
- After thawing components on ice, prepare 10 μl cap methyltransferase reactions in 1.7 ml tubes on ice by combining the following in order: RNase-free water (for 10 μl total volume), 1 μl of 10x cap methylation buffer (see Recipes), 1 μl of 1 μM SAM, 1 μl of 20 mM DTT, 1 μl of RNaseOUT, nuclear or cytoplasmic extract for 1 μg of total protein, 5 fmol of [32P]G-capped RNA. Using a master mix of the reaction components (omitting the extracts) is recommended if working with a large number of extract samples. Gently mix, incubate at 37 °C for 30 min, and then immediately return to ice.
Data analysis
The extent of conversion of GpppRNA to m7GpppRNA during a cap methyltransferase reaction (and by extension, the cap methyltransferase activity present in the corresponding biological sample) is represented by the ratio of the m7GpppG spot intensity to the sum of the GpppG and m7GpppG spot intensities on the TLC plate phosphor image. The spot intensities can be determined using any image software that includes densitometry analysis, such as ImageQuant TL. To ensure consistent calculation of spot intensities for multiple samples, use lanes or boxes of the same size that are large enough to just fit the entire spot for densitometry analysis. Also ensure that any background signal is subtracted; in ImageQuant TL, we use the ‘rubberband’ setting for establishing the baseline for peak signal integration. To account for biological variability among samples when comparing multiple conditions, it is good practice to perform this assay with at least three biological replicates to enable appropriate statistical testing.
Notes
- The recombinant human capping enzyme we used to generate the radiolabeled GpppRNA was produced in-house as described in Trotman et al. (2017). Despite this enzyme stock having robust capping activity with non-radiolabeled GTP, the production of radiolabeled GpppRNA was rather inefficient. If a recombinant capping enzyme is not available, we recommend using a commercially available enzyme, such as the Vaccinia Capping Enzyme (NEB, catalog #M2080S), to produce the GpppRNA as was used in previous reports (Pillutla et al., 1998; Cowling, 2010). If Vaccinia Capping Enzyme is used to generate GpppRNA, be sure to omit SAM from this reaction, as this enzyme also contains cap methyltransferase activity.
- Unlike most types of chromatographic systems, the flow rate of TLC is variable and cannot be easily adjusted to optimize performance according to the van Deemter Equation (van Deemter et al., 1956; Guiochon et al., 1979). In TLC, the capillary flow rate is dependent on properties such as the particle size of the TLC plate, which can vary substantially from manufacturer to manufacturer. We have thus found that some manufacturers’ PEI cellulose plates give streakier, more poorly resolved results than others, with the Macherey-Nagel plates listed here giving the best performance of those we have tested under the given conditions.
- Any short, in-vitro-transcribed RNA (with a 5’ triphosphate) should be suitable for generating the radiolabeled GpppRNA substrate, and this protocol can be modified to produce other GpppRNA substrates if desired. The protocol presented here simply describes an easy way to generate the same 32 nt substrate RNA used in the initial characterization of human RNMT (Pillutla et al., 1998) and in our recent study (Trotman et al., 2017).
- Use fresh [α-32P]GTP to maximize the ratio of radioactive-to-nonradioactive GTP in the stock, which will allow for greater signal intensity during phosphorimaging. To account for radioactive decay of the [α-32P]GTP stock, we recommend using the PerkinElmer radioactivity calculator (https://www.perkinelmer.com/tools/calculatorrad) to determine the concentration of total GTP on the day of its use.
- As an additional control for visualizing the position of m7GpppG and ensuring success of the assay, we suggest preparing a parallel cap methyltransferase activity reaction using a recombinant cap methyltransferase (such as Vaccinia Capping Enzyme from NEB).
- The radioisotope 32P emits beta particles that can be potentially harmful if not handled properly. To ensure safety while handling 32P, always wear eye and skin protection, work behind an acrylic bench shield, and use a Geiger counter to frequently check surfaces for contamination. We recommend using a dedicated set of pipets for use with radioactive materials in case they become contaminated. Store all radioactive samples at 4 °C or -20 °C in secondary acrylic storage boxes. Clean up any spills, no matter how small, and notify lab personnel and appropriate safety officials in the case of a sufficiently large spill. All radioactive waste should be placed in acrylic waste bins for at least 10 half-lives (about 143 days) to decay in storage. Contact your institution’s radiation safety office for additional information.
Recipes
- 1 M DTT
Dissolve 1.54 g of DTT in RNase-free water to a total volume of 10 ml. Aliquot and store at -20 °C - 1x TBE (running buffer for urea-PAGE)
Bring 100 ml of RNase-free 10x TBE (Reagent #47) to 1 L with RNase-free water. Store at room temp. - 20 mM DTT
Bring 20 μl of 1 M DTT to 1 ml with RNase-free water. Aliquot and store at -20 °C - 100 mM PMSF in isopropanol
Dissolve 1.74 g of PMSF in isopropanol to a total volume of 10 ml. Aliquot and store at -20 °C - 10x Tris/glycine
Dissolve 30 g of Tris and 144 g of glycine in RNase-free water to a total volume of 1 l. Store at room temp. - 10% (w/v) SDS
Dissolve 10 g of SDS in RNase-free water to a total volume of 100 ml. Store at room temp. - Tris/glycine/SDS running buffer for SDS-PAGE
890 ml RNase-free water
100 ml 10x Tris/glycine
10 ml 10% (w/v) SDS - Tris/glycine/methanol/SDS transfer buffer
To 200 ml of methanol, add 500 ml of RNase-free water and then 100 ml of 10x Tris/glycine and 1 ml of 10% (w/v) SDS. Add RNase-free water to a total volume of 1 L and store at 4 °C - 20x Tris-buffered saline (TBS)
Dissolve 48.4 g of Tris and 160 g of NaCl in 800 ml of RNase-free water and adjust the pH to 7.6 with concentrated HCl. Bring the total volume to 1 L with RNase-free water and store at room temp. - 3% BSA in TBS
To 1.5 g of BSA, add 2.5 ml of 20x TBS, and dissolve in RNase-free water to a total volume of 50 ml. Store at 4 °C - 40% (v/v) Tween 20
Mix 20 ml Tween 20 with RNase-free water to a total volume of 50 ml. Store at room temp. - TBS-T
474 ml RNase-free water
25 ml 20x TBS
1.25 ml 40% (v/v) Tween 20 - 1 μM SAM
Dilute 32 mM SAM 320-fold with RNase-free water to produce a 100 μM stock, and then dilute this 100 μM stock 100-fold with RNase-free water. Aliquot and store at -20 °C - 500 mM sodium acetate, pH 5.2
Dilute 3 M sodium acetate, pH 5.2 6-fold with RNase-free water. Store at room temp. - 0.4 M ammonium sulfate
Dissolve 52.86 g of ammonium sulfate in RNase-free water to 1 L. Store at room temp. - 10% IGEPAL CA-630 (v/v) in water
Add RNase-free water to 1 ml IGEPAL CA-630 to a total volume of 10 ml. Store at room temp. - 10x annealing buffer
100 mM Tris-HCl pH 7.5
500 mM NaCl - 4x capping buffer
40 mM Tris pH 7.5
12 mM MgCl2
4 mM DTT
0.4 mM EDTA
80% (v/v) glycerol - YO Lysis Buffer
10 mM HEPES pH 7.3
10 mM KCl
10 mM MgCl2
0.2% (v/v) IGEPAL CA-630
2 mM DTT*
0.5 mM PMSF*
7.5 μl/ml protease inhibitor cocktail (Sigma-Aldrich)*
7.5 μl/ml phosphatase inhibitor cocktail 2 (Sigma-Aldrich)*
7.5 μl/ml phosphatase inhibitor cocktail 3 (Sigma-Aldrich)* - YO Buffer A
10 mM HEPES pH 7.3
25% (v/v) glycerol
420 mM NaCl
1.5 mM MgCl2
0.2 mM EDTA
1 mM DTT*
0.5 mM PMSF*
7.5 μl/ml protease inhibitor cocktail (Sigma-Aldrich)*
7.5 μl/ml phosphatase inhibitor cocktail 2 (Sigma-Aldrich)*
7.5 μl/ml phosphatase inhibitor cocktail 3 (Sigma-Aldrich)*
- 10x cap methylation buffer
500 mM Tris pH 8
60 mM KCl
12.5 mM MgCl2
Acknowledgments
This work was supported by R01 grant GM084177 from the National Institutes of Health (to D.R.S). J.B.T. was funded by NIH T32 training grant GM08512 (to The Ohio State University) and by a pre-doctoral fellowship from The Ohio State University Center for RNA Biology. The protocol presented here was adapted from those in Pillutla et al., 1998, Otsuka et al., 2009, and Cowling, 2010 and was used for experiments in Trotman et al., 2017. The authors declare no conflicts of interest or competing interests.
References
- Aouadi, W., Eydoux, C., Coutard, B., Martin, B., Debart, F., Vasseur, J. J., Contreras, J. M., Morice, C., Querat, G., Jung, M. L., Canard, B., Guillemot, J. C. and Decroly, E. (2017). Toward the identification of viral cap-methyltransferase inhibitors by fluorescence screening assay. Antiviral Res 144: 330-339.
- Cowling, V. H. (2010). Enhanced mRNA cap methylation increases cyclin D1 expression and promotes cell transformation. Oncogene 29(6): 930-936.
- Gonatopoulos-Pournatzis, T., Dunn, S., Bounds, R. and Cowling, V. H. (2011). RAM/Fam103a1 is required for mRNA cap methylation. Mol Cell 44(4): 585-596.
- Guiochon, G., Bressolle, F. and Siouffi, A. (1979). Study of the performances of thin layer chromatography IV – Optimization of experimental conditions. J Chromatogr Sci 17: 368-386.
- Jiao, X., Chang, J. H., Kilic, T., Tong, L. and Kiledjian, M. (2013). A mammalian pre-mRNA 5' end capping quality control mechanism and an unexpected link of capping to pre-mRNA processing. Mol Cell 50(1): 104-115.
- Niedzwiecka, A., Marcotrigiano, J., Stepinski, J., Jankowska-Anyszka, M., Wyslouch-Cieszynska, A., Dadlez, M., Gingras, A. C., Mak, P., Darzynkiewicz, E., Sonenberg, N., Burley, S. K. and Stolarski, R. (2002). Biophysical studies of eIF4E cap-binding protein: recognition of mRNA 5’ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J Mol Biol 319(3): 615-635.
- Otsuka, Y., Kedersha, N. L. and Schoenberg, D. R. (2009). Identification of a cytoplasmic complex that adds a cap onto 5'-monophosphate RNA. Mol Cell Biol 29(8): 2155-2167.
- Peyrane, F., Selisko, B., Decroly, E., Vasseur, J. J., Benarroch, D., Canard, B. and Alvarez, K. (2007). High-yield production of short GpppA- and 7MeGpppA-capped RNAs and HPLC-monitoring of methyltransfer reactions at the guanine-N7 and adenosine-2'O positions. Nucleic Acids Res 35(4): e26.
- Pillutla, R. C., Yue, Z., Maldonado, E. and Shatkin, A. J. (1998). Recombinant human mRNA cap methyltransferase binds capping enzyme/RNA polymerase IIo complexes. J Biol Chem 273(34): 21443-21446.
- Trotman, J. B., Giltmier, A. J., Mukherjee, C. and Schoenberg, D. R. (2017). RNA guanine-7 methyltransferase catalyzes the methylation of cytoplasmically recapped RNAs. Nucleic Acids Res 45(18): 10726-10739.
- van Deemter, J. J., Zuiderweg, F. J. and Klinkenberg, A. (1956). Longitudinal diffusion and resistance to mass transfer as causes of nonideality in chromatography. Chem Eng Sci 5: 271-289.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Trotman, J. and Schoenberg, D. R. (2018). RNA Cap Methyltransferase Activity Assay. Bio-protocol 8(6): e2767. DOI: 10.21769/BioProtoc.2767.
Category
Molecular Biology > RNA > RNA capping
Biochemistry > Other compound > Nucleoside triphosphate
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











