- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Synthetic Genetic Interaction (CRISPR-SGI) Profiling in Caenorhabditis elegans
Published: Vol 8, Iss 5, Mar 5, 2018 DOI: 10.21769/BioProtoc.2756 Views: 8734
Reviewed by: Gal HaimovichManish ChamoliAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Editing the Serratia proteamaculans Genome Using the Allelic Exchange Method
Ksenia Chukhontseva [...] Ilya Demidyuk
Sep 20, 2025 1387 Views

Creating Loss-of-Function Mutants of Neurospora crassa Using a Novel CRISPR/Cas9 System
Stefanie Grüttner
Jan 5, 2026 199 Views

Identifying Causal Genes and Building Regulatory Networks in Crops Using the CisTrans-ECAS Method
Yutong Yan [...] Weibo Xie
Feb 5, 2026 97 Views
Abstract
Genetic interaction screens are a powerful methodology to establish novel roles for genes and elucidate functional connections between genes. Such studies have been performed to great effect in single-cell organisms such as yeast and E. coli (Schuldiner et al., 2005; Butland et al., 2008; Costanzo et al., 2010), but similar large-scale interaction studies using targeted reverse-genetic deletions in multi-cellular organisms have not been feasible. We developed a CRISPR/Cas9-based method for deleting genes in C. elegans and replacing them with a heterologous fluorescent reporter (Norris et al., 2015). Recently we took advantage of that system to perform a large-scale, reverse genetic screen using null alleles in animals for the first time, focusing on RNA binding protein genes (Norris et al., 2017). This type of approach should be similarly applicable to many other gene classes in C. elegans. Here we detail the protocols involved in generating a library of double mutants and performing medium-throughput competitive fitness assays to test for genetic interactions resulting in fitness changes.
Keywords: C. elegansBackground
Large-scale genetic interaction screens using reverse-genetic null alleles have not previously been feasible in animals. RNAi has been used to study genetic interactions in C. elegans by knocking down expression of a large number of different genes in the presence of a single mutant background (Baugh et al., 2005; Lehner et al., 2006). However, this strategy is limited by the variable efficacy of RNAi knockdown, thereby complicating the interpretation of the results. We developed a method for efficient editing of the C. elegans genome (Norris et al., 2015) and recently expanded upon that method to enable large-scale genetic interaction profiling in animals using null alleles for the first time. We focused our initial efforts on neuronally-expressed RNA binding protein genes, which have been shown in a number of cases to act combinatorially (Gracida et al., 2016; Norris et al., 2014). We found widespread genetic interactions among the set of RNA binding proteins we studied, and similar strategies should be broadly applicable to other gene classes as well.
Materials and Reagents
- Platinum Wire for worm pick, 30 gauge 0.254 mm diameter (e.g., Genesee Scientific, catalog number: 59-30P6 )
- Worm pick handle (e.g., Genesee Scientific, catalog number: 59-AWP )
- 6 cm Petri dishes (e.g., Fisher Scientific, catalog number: FB0875713A)
Manufacturer: Corning, catalog number: 431762 .
- Wild-type (N2) male and hermaphrodite worms (available from CGC, strain N2)
- Monobasic potassium phosphate (KH2PO4) (e.g., Sigma-Aldrich, catalog number: 1551139 )
- Solid KOH
- Sodium chloride (NaCl) (e.g., Genesee Scientific, catalog number: 18-214 )
- Agar (e.g., Genesee Scientific, catalog number: 20-249 )
- peptone (e.g., Genesee Scientific, catalog number: 20-261 )
- Cholesterol (Sigma-Aldrich, catalog number: C8667 )
- 95% ethanol (e.g., Sigma-Aldrich, catalog number: 792799 )
- Calcium chloride (CaCl2) (e.g., Sigma-Aldrich, catalog number: C1016 )
- Magnesium sulfate (MgSO4) (e.g., Sigma-Aldrich, catalog number: M7506 )
- 1 M potassium phosphate (pH 6)
- Nematode Growth Media (NGM) (see Recipes)
Equipment
- Fluorescent dissecting stereomicroscope with high sensitivity for single-copy fluorescence detection (e.g., ZEISS, model: Axio Zoom.V16 or Leica, model: Leica M165 FC )
- 25 °C incubator (e.g., VWR, Sheldon Manufacturing, model: Model 2005 )
- Autoclave
Procedure
- Creation of double mutants
The generation of single mutants using CRISPR/Cas9 has been covered elsewhere (Norris et al., 2015). This protocol begins with two single mutant worms in which the genes of interest have been deleted and replaced by compatible heterologous fluorescent reporters. In this example, the reporters are myo-2::GFP (expressed in pharyngeal muscles) and myo-3::GFP (expressed in body wall muscles).- To create a male strain necessary for crossing, there are two options:
- Cross ~5 wild-type (N2) male worms (available from CGC) with ~5 hermaphrodites of mutant strain #1 to create male progeny that are heterozygous for the mutation (verify by ensuring the males are fluorescent).
- Obtain homozygous males of mutant strain #1 according to standard protocols (He, 2011a).
- Cross ~5 wild-type (N2) male worms (available from CGC) with ~5 hermaphrodites of mutant strain #1 to create male progeny that are heterozygous for the mutation (verify by ensuring the males are fluorescent).
- Cross males from mutant strain #1 with hermaphrodites from mutant strain #2 (see Figure 1).
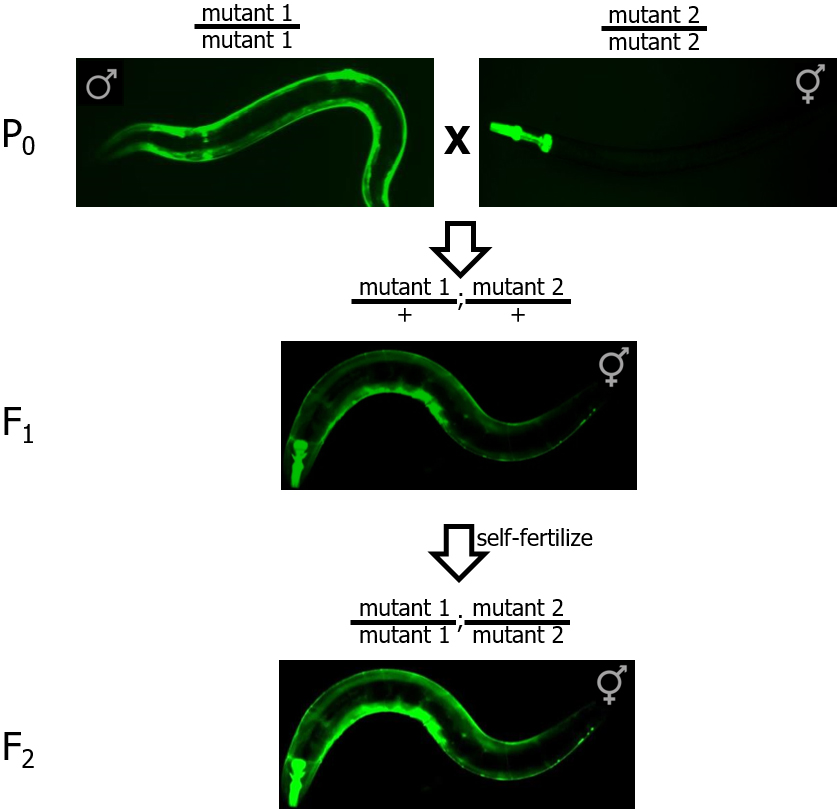
Figure 1. Crossing strategy to create double mutant worms from individual single mutant strains. GFP is used to distinguish homozygous mutants (P0 and F2 panels, brighter fluorescence) from heterozygous mutants (F1 panel, dimmer fluorescence).
- Pick a single cross-progeny hermaphrodite containing both GFP markers (both myo-2::GFP and myo-3::GFP) onto a new plate. These worms correspond to double heterozygotes (i.e., mutant1/+; mutant2/+). Allow these hermaphrodites to self-fertilize.
- To obtain double-homozygous strains, pick worms with brighter GFP expression coming from both promoters. On a good-quality fluorescence dissection scope, you should be able to tell a difference in brightness between a strain with one copy (heterozygous mutant) and two copies (homozygous mutant) of the GFP transgene (see Figure 2).
Note: Alternatively, pick a larger number of worms to individual plates and determine double homozygotes as worms with progeny that are 100% fluorescent for both transgenes.
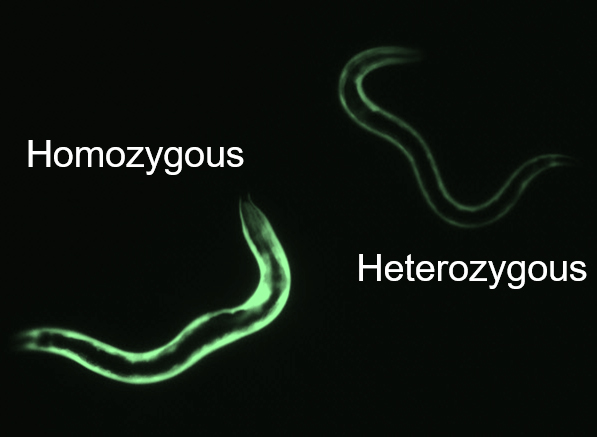
Figure 2. Fluorescent brightness reveals mutant genotypes. Worms imaged in situ on NGM plates demonstrating that homozygous mutants (left) are noticeably brighter than heterozygous mutants (right).
- To create a male strain necessary for crossing, there are two options:
- Competitive fitness assay
The competitive fitness assay (Figure 3A) provides a simple and relatively high-throughput method for assessing phenotypes of large numbers of mutants and can be performed immediately after strain generation.- Pick equal numbers of wild-type (N2) and mutant worms onto one seeded NGM plate (see Recipes).
Note: We find that 4 staged L4s of each genotype (i.e., 4 wild-type L4s and 4 mutant L4s) on a 6 cm plate works best to support 2 generations of growth without starving out the plate.
- Incubate plate at 25 °C for 5 days.
- Under a fluorescent stereomicroscope count the number of fluorescent (mutant) worms versus non-fluorescent (wild-type) worms. We prefer to count from pre-defined locations on each plate (e.g., 50 worms from the middle of the plate, 50 from the periphery, and 50 from in between).
Notes:- We recommend counting all hatched worms regardless of stage.
- For ease of counting, worms can be treated with anesthetic or cooled at 4 °C for an hour or longer.
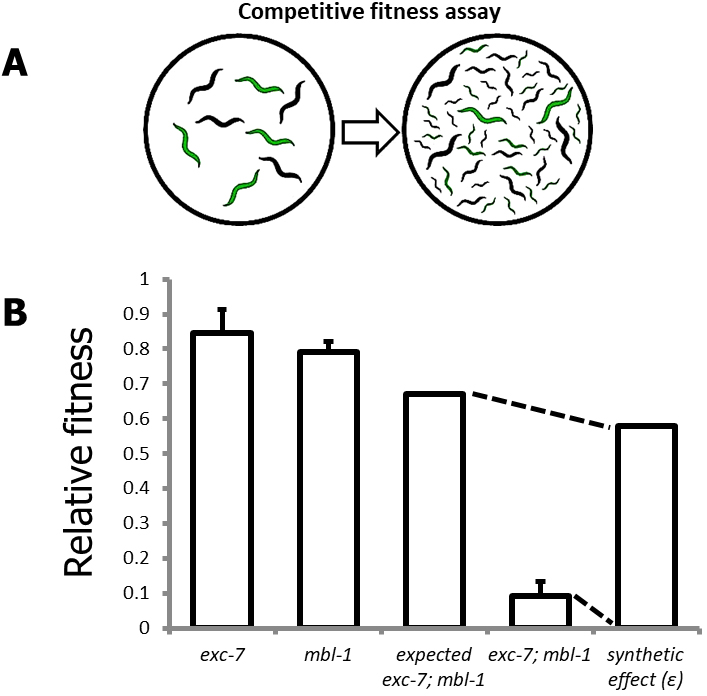
Figure 3. The competitive fitness assay. A. Schematic overview of competitive fitness assays; B. Representative visualization of synthetic effect (ɛ) calculation for a genetic interaction between the genes exc-7 and mbl-1.
- We recommend counting all hatched worms regardless of stage.
- Pick equal numbers of wild-type (N2) and mutant worms onto one seeded NGM plate (see Recipes).
Data analysis
These data analysis steps are an expansion of those detailed in Norris et al. (2017).
- For robustness, 3 biological replicates should be performed, each on a different day and different plate.
- Relative fitness scores can be calculated by the following formula:
% mutant/% expected (i.e., 50%) = relative fitness
This will yield a relative fitness value ranging from 0 (strong loss of fitness) to 2 (strong increase in fitness), with a value of 1 indicating no change of fitness compared to wild-type.
- To generate an expected fitness value (see Figure 3B for representative example) for a double mutant (Fexp1,2) based on the null hypothesis of no genetic interaction between mutant 1 (F1) and mutant 2 (F2), use the following formula (Mani et al., 2008; Baryshnikova et al., 2010):
Fexp1,2 = F1 x F2
- To compare the observed fitness of the double mutant (Fobs1,2) to the expected fitness values:
ɛ = Fobs1,2 - Fexp1,2
This yields the ‘synthetic effect’ score (ɛ). Negative ɛ values indicate a strain with lower fitness than expected, and positive ɛ values indicate a strain with higher fitness than expected.
- To report statistically significant results, we set a conservative threshold of |ɛ| ≥ 0.20. For strains passing the |ɛ| ≥ 0.20 threshold, a Fisher’s exact test is applied to the aggregate observed values and the null-expectation values with a Bonferroni-corrected P-value of < 0.01 used as the significance threshold.
Notes
- Ensure that worm strains are treated identically for at least 3 days before they are picked onto competition assay plates (e.g., healthy, unstarved, uncrowded plates grown at the same temperature).
- The competition assay can be adapted to a variety of different types of mutants and conditions; the only requirement is that the two strains to be compared have some easily-distinguishable feature (e.g., fluorescence)
Recipes
- 1 M potassium phosphate (pH 6) (1 L)
Dissolve 136.1 g KH2PO4 in about 800 ml dH2O
Adjust pH to 6.0 with solid KOH (approx. 15 g) before bringing up to volume
Make 100 ml aliquots and autoclave
- Nematode Growth Media (NGM)
Note: Recipe from ‘Common Worm Media and Buffers’ (He, 2011b).
For 1 L medium
3 g NaCl
17 g agar
2.5 g peptone
1 ml cholesterol (5 mg ml-1 in 95% EtOH)
975 ml ddH2O
Autoclave, and then add the following sterile solution (autoclaved)
1 ml 1 M CaCl2
1 ml 1 M MgSO4
25 ml 1 M potassium phosphate (pH 6) (to avoid precipitation, mix between addition of MgSO4 and potassium phosphate)
Typically pour 60 x 6 mm plate (~10 ml of media per plate) and store NGM plates in plastic boxes with covers at room temperature
Acknowledgments
This protocol was adapted from Norris et al., 2017. Support for AN was supplied by the Floyd B. James Endowed Professorship (Southern Methodist University). Support for JC was supplied by NIH Office of the Director (NIH Early Independence Award DP5OD009153) and Natural Sciences and Engineering Research Council of Canada (Discovery Grant RGPIN-2017-06573). There are no conflicts of interest or competing interest.
References
- Baryshnikova, A., Costanzo, M., Kim, Y., Ding, H., Koh, J., Toufighi, K., Youn, J. Y., Ou, J., San Luis, B. J., Bandyopadhyay, S., Hibbs, M., Hess, D., Gingras, A. C., Bader, G. D., Troyanskaya, O. G., Brown, G. W., Andrews, B., Boone, C. and Myers, C. L. (2010). Quantitative analysis of fitness and genetic interactions in yeast on a genome scale. Nat Methods 7(12): 1017-1024.
- Baugh, L. R., Wen, J. C., Hill, A. A., Slonim, D. K., Brown, E. L. and Hunter, C. P. (2005). Synthetic lethal analysis of Caenorhabditis elegans posterior embryonic patterning genes identifies conserved genetic interactions. Genome Biol 6(5): R45.
- Butland, G., Babu, M., Diaz-Mejia, J. J., Bohdana, F., Phanse, S., Gold, B., Yang, W., Li, J., Gagarinova, A. G., Pogoutse, O., Mori, H., Wanner, B. L., Lo, H., Wasniewski, J., Christopolous, C., Ali, M., Venn, P., Safavi-Naini, A., Sourour, N., Caron, S., Choi, J. Y., Laigle, L., Nazarians-Armavil, A., Deshpande, A., Joe, S., Datsenko, K. A., Yamamoto, N., Andrews, B. J., Boone, C., Ding, H., Sheikh, B., Moreno-Hagelseib, G., Greenblatt, J. F. and Emili, A. (2008). eSGA: E. coli synthetic genetic array analysis. Nat Methods 5(9): 789-795.
- Costanzo, M., Baryshnikova, A., Bellay, J., Kim, Y., Spear, E. D., Sevier, C. S., Ding, H., Koh, J. L., Toufighi, K., Mostafavi, S., Prinz, J., St Onge, R. P., VanderSluis, B., Makhnevych, T., Vizeacoumar, F. J., Alizadeh, S., Bahr, S., Brost, R. L., Chen, Y., Cokol, M., Deshpande, R., Li, Z., Lin, Z. Y., Liang, W., Marback, M., Paw, J., San Luis, B. J., Shuteriqi, E., Tong, A. H., van Dyk, N., Wallace, I. M., Whitney, J. A., Weirauch, M. T., Zhong, G., Zhu, H., Houry, W. A., Brudno, M., Ragibizadeh, S., Papp, B., Pal, C., Roth, F. P., Giaever, G., Nislow, C., Troyanskaya, O. G., Bussey, H., Bader, G. D., Gingras, A. C., Morris, Q. D., Kim, P. M., Kaiser, C. A., Myers, C. L., Andrews, B. J. and Boone, C. (2010). The genetic landscape of a cell. Science 327(5964): 425-431.
- Gracida, X., Norris, A. D. and Calarco, J. A. (2016). Regulation of tissue-specific alternative splicing: C. elegans as a model system. Adv Exp Med Biol 907: 229-261.
- He, F. (2011a). Making males of C. elegans. Bio-protocol e58.
- He, F. (2011b). Common worm media and buffers. Bio-protocol e55.
- Lehner, B., Crombie, C., Tischler, J., Fortunato, A. and Fraser, A. G. (2006). Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat Genet 38(8): 896-903.
- Mani, R., St Onge, R. P., Hartman, J. L., Giaever, G. and Roth, F. P. (2008). Defining genetic interaction. Proc Natl Acad Sci U S A 105: 3461-3466.
- Norris, A. D., Gao, S., Norris, M. L., Ray, D., Ramani, A. K., Fraser, A. G., Morris, Q., Hughes, T. R., Zhen, M. and Calarco, J. A. (2014). A pair of RNA-binding proteins controls networks of splicing events contributing to specialization of neural cell types. Mol Cell 54(6): 946-959.
- Norris, A. D., Gracida, X. and Calarco, J. A. (2017). CRISPR-mediated genetic interaction profiling identifies RNA binding proteins controlling metazoan fitness. Elife 6.
- Norris, A. D., Kim, H. M., Colaiacovo, M. P. and Calarco, J. A. (2015). Efficient genome editing in Caenorhabditis elegans with a toolkit of dual-marker selection cassettes. Genetics 201(2): 449-458.
- Schuldiner, M., Collins, S. R., Thompson, N. J., Denic, V., Bhamidipati, A., Punna, T., Ihmels, J., Andrews, B., Boone, C., Greenblatt, J. F., Weissman, J. S. and Krogan, N. J. (2005). Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 123(3): 507-519.
Article Information
Copyright
Calarco and Norris. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Calarco, J. A. and Norris, A. D. (2018). Synthetic Genetic Interaction (CRISPR-SGI) Profiling in Caenorhabditis elegans. Bio-protocol 8(5): e2756. DOI: 10.21769/BioProtoc.2756.
- Norris, A. D., Gracida, X. and Calarco, J. A. (2017). CRISPR-mediated genetic interaction profiling identifies RNA binding proteins controlling metazoan fitness. Elife 6.
Category
Systems Biology > Interactome > Gene network
Molecular Biology > DNA > Mutagenesis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








