- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Flow Cytometric Quantification of Fatty Acid Uptake by Mycobacterium tuberculosis in Macrophages
Published: Vol 8, Iss 4, Feb 20, 2018 DOI: 10.21769/BioProtoc.2734 Views: 12817
Reviewed by: Longping Victor TseShalini Low-NamAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In vitro Assessment of Efferocytic Capacity of Human Macrophages Using Flow Cytometry
Ana C.G. Salina [...] Larissa D. Cunha
Dec 20, 2023 5155 Views

Quantification of Macrophage Cellular Ferrous Iron (Fe2+) Content using a Highly Specific Fluorescent Probe in a Plate-Reader
Philipp Grubwieser [...] Christa Pfeifhofer-Obermair
Feb 5, 2024 2215 Views

Novel Experimental Approach to Investigate Immune Control of Vascular Function: Co-culture of Murine Aortas With T Lymphocytes or Macrophages
Taylor C. Kress [...] Eric J. Belin de Chantemèle
Sep 5, 2025 3547 Views
Abstract
Mycobacterium tuberculosis (Mtb) has evolved to assimilate fatty acids from its host. However, until recently, there was no reliable way to quantify fatty acid uptake by the bacteria during host cell infection. Here we describe a new method to quantify fatty acid uptake by intracellular bacilli. We infect macrophages with Mtb constitutively expressing mCherry and then metabolically label them with Bodipy-palmitate. Following the labeling procedure, we isolate Mtb-containing phagosomes on a sucrose cushion and disrupt the phagosomes with detergent. After extensive washes, the isolated bacteria are analyzed by flow cytometry to determine the level of Bodipy-palmitate signal associated with the bacteria. Using a Mtb mutant strain defective in fatty acid uptake in liquid culture we determined that this mutant assimilated 10-fold less Bodipy-palmitate than the wild type strain during infection in macrophages. This quantitative method of fatty acid uptake can be used to further identify pathways involved in lipid uptake by intracellular Mtb and possibly other bacteria.
Keywords: Fatty acidBackground
The ability of Mycobacterium tuberculosis (Mtb) to assimilate host-derived lipids (fatty acids and cholesterol) enables survival of the pathogen within its host (Russell et al., 2010; Lovewell et al., 2016). This idea is supported by upregulation of cholesterol and fatty acid metabolism-related genes by Mtb inside of macrophage, during mouse infection and in human lung tissue (Schnappinger et al., 2003; Rachman et al., 2006; Rohde et al., 2007; Fontán et al., 2008; Tailleux et al., 2008; Homolka et al., 2010; Rohde et al., 2012). The importance of cholesterol metabolism for Mtb during infection is supported by genetic studies and by identification of new antituberculosis compounds targeting cholesterol metabolism (Pandey and Sassetti, 2008; Wipperman et al., 2014; VanderVen et al., 2015). However, discovery of the specific machinery devoted to fatty acid uptake in Mtb is hindered not only by apparent redundancy of dedicated genes (Cole et al., 1998) but also by a paucity of reliable assays. Metabolic labeling with radioactive substrate has been the accepted approach to assess efficiency of fatty acid intake by bacterium in broth culture (Forrellad et al., 2014). This method is extremely challenging to apply for intracellular Mtb, and few groups have reported successful use of this approach (Daniel et al., 2011). Alternatively, TEM and staining with lipophilic dyes such as Bodipy 493/503, Nile Red, Oil Red can facilitate detection of lipids inside of mycobacteria during infection (Daniel et al., 2011; Podinovskaia et al., 2013; Caire-Brändli et al., 2014). However, neither of these labeling approaches directly assess the active import of substrate, but instead indicate the total amount of accumulated lipids. Therefore, a means of metabolic labeling of active fatty acid import by Mtb during infection that is tractable to downstream characterization is sorely needed.
Recently, it was shown that fluorescent fatty acids can be delivered effectively to intracellular bacteria and can be detected by microscopy (Podinovskaia et al., 2013). These observations led us to develop a new method of flow-cytometry based quantification of fluorescent fatty acid uptake by Mtb within its host cell. This assay allowed us to demonstrate that a ∆lucA::hyg Mtb strain is defective in fatty acid uptake during macrophage infection (Nazarova et al., 2017) (Figure 1). We believe that this methodology opens the door to genetic screens to further understand mechanisms involved in fatty acid uptake by Mtb and possibly other intracellular pathogens during infection in host cells.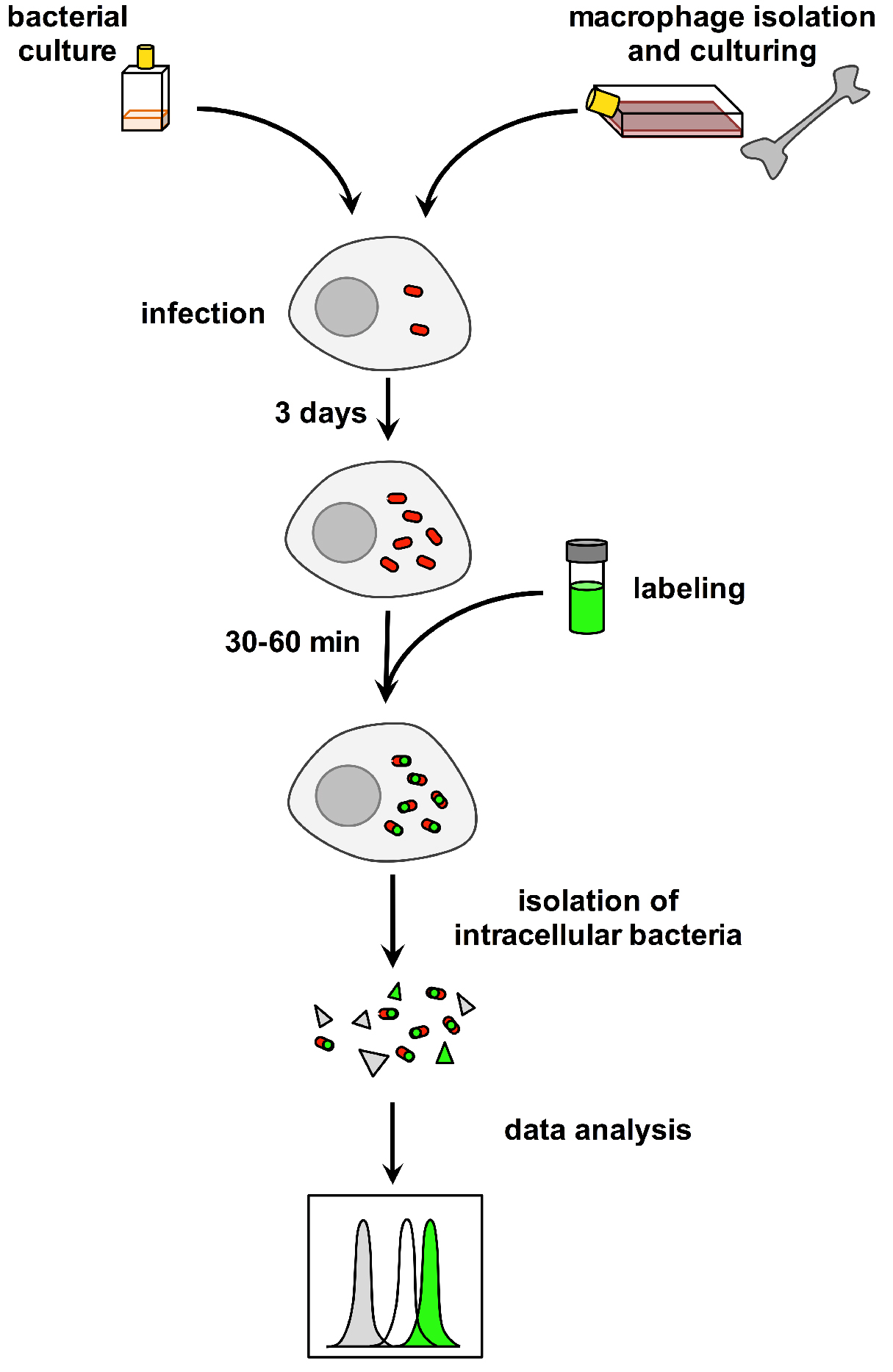
Figure 1. Overview of the method used to quantify fatty acid uptake by M. tuberculosis during infection in macrophages
Materials and Reagents
- Serological pipets, 5 ml, 10 ml, 25 ml and 50 ml (Corning, Costar®, catalog numbers: 4487 , 4488 , 4489 and 4490 )
- Pipette tips, 20 μl, 100 μl, 200 μl, 1 ml (Biotix, Neptune®, catalog numbers: BT20 , BT100 , BT200 ; Thermo Fisher Scientific, Thermo Scientific, catalog number: 2079-HR )
- T25 (sterile 25 cm2 tissue culture flasks with filtered cap) (TPP, catalog number: 90026 )
- T75 or T150 (sterile 75 cm2 or 150 cm2 tissue culture flasks with filtered cap) (TPP, catalog numbers: 90076 or 90151 )
- Sterile 1 ml tuberculin syringe with 25 gauge needle (BD, catalog number: 309626 )
- Cell scrapers, 25 cm (SARSTEDT, catalog number: 83.1830 )
- 15 ml conical tube (SARSTEDT, catalog number: 62.554.100 )
- 50 ml conical tube (SARSTEDT, catalog number: 62.547.100 )
- Glasstic® slides with grids (KOVA International, catalog number: 87144 )
- 150 x 15 mm Petri dish (VWR, catalog number: 25384-326 )
- 2 ml screw-cap tubes (VWR, catalog number: 16466-042 )
- FACS tubes (VWR, catalog number: 60818-496 )
- 3 ml syringe (with Luer-LokTM tip, BD, catalog number: 309657 )
- Disposable plastic OD cuvettes with square caps (Fisher Scientific, FisherbrandTM catalog numbers: 14-955-128 and 14-385-999 )
- Mycobacterium tuberculosis constitutively expressing mCherry (pMV306 smyc’::mCherry (Kanr)) Source: Russell and VanderVen lab (Nazarova et al., 2017)
- Bone marrow-derived murine macrophages (BALB/c mice, THE JACKSON LABORATORIES, catalog number: 000651 )
Note: Differentiation is described in detail in Nazarova et al. (2017). - L cells (NCTC clone 929 [L cell, L-929, derivative of Strain L]) (ATCC, catalog number: CCL-1 )
- Phosphate-buffered saline (PBS) 1x (Mediatech, catalog number: 21-040 )
- Tyloxapol (Acros Organics, catalog number: 422370050 )
- Middlebrook 7H9 Broth Base (BD, DifcoTM, catalog number: 271310 )
- Distilled water
- Glycerol (VWR, catalog number: 97062-452 )
- Middlebrook OADC Enrichment (BD, BBLTM, catalog number: 212351 )
- Kanamycin sulfate (IBI Scientific, catalog number: IB02120 )
- Heat inactivated fetal bovine serum (Thermo Fisher Scientific, GibcoTM, catalog number: 10437028 )
Note: Heat inactivated at 56 °C for 30 min. - 200 mM L-glutamine (100x) (Mediatech, catalog number: 25-005 )
- 100 mM sodium pyruvate (Mediatech, catalog number: 25-000 )
- Penicillin-streptomycin solution 100x (Mediatech, catalog number: 30-002 )
- Dulbecco’s modification of Eagle’s medium (DMEM) 1x (Mediatech, catalog number: 10-017 )
- Glucose (dextrose) (Fisher Scientific, catalog number: BP350-1 )
- Bovine serum albumin (BSA) (Roche Diagnostics, catalog number: 03116964001 )
- Gelatin from cold water fish skin (Sigma-Aldrich, catalog number: G7765 )
- Calcium chloride (CaCl2) (Sigma-Aldrich, catalog number: C4901 )
- Magnesium chloride (MgCl2) (AMRESCO, catalog number: J364 )
- Fatty acid-free BSA (Roche Diagnostics, catalog number: 03117057001 )
- 100% ethanol, 200 Proof (Decon Labs, catalog number: V1016 )
- BodipyTM FL C16 (Bodipy-palmitate) (Thermo Fisher Scientific, InvitrogenTM, catalog number: D3821 )
- BodipyTM FL C12 (Thermo Fisher Scientific, InvitrogenTM, catalog number: D3822 )
- BodipyTM 558/568 C12 (Thermo Fisher Scientific, InvitrogenTM, catalog number: D3835 )
- Potassium chloride (KCl) (Mallinckrodt Chemicals or Avantor Performance Materials, MACRON, catalog number: 6858-04 )
- Sucrose (Avantor Performance Materials, J.T. Baker, catalog number: 4097 )
- Ethylenebis(oxyethylenenitrilo)tetraacetic acid (EGTA) (Avantor Performance Materials, J.T. Baker, catalog number: L657 )
- HEPES (VWR, catalog number: BDH4162 )
- TweenTM 80 (Fisher Chemical, catalog number: T164-500 )
- Paraformaldehyde (PFA) (Sigma-Aldrich, catalog number: 158127 )
- 20% tyloxapol (see Recipes)
- 7H9 OADC media(see Recipes)
- Kanamycin 25 mg/ml (see Recipes)
- D10 media (see Recipes)
- L-cell conditioned media (see Recipes)
- BMDM media (see Recipes)
- Basal uptake buffer (BUB) (see Recipes)
- 1% fatty acid-free BSA (see Recipes)
- 4 mM Bodipy-palmitate (see Recipes)
- Cuvette buffer (see Recipes)
- Homogenization buffer (see Recipes)
- 20% Tween 80 (see Recipes)
- 0.05% tyloxapol (see Recipes)
- 4% PFA (see Recipes)
Equipment
- Pipette controller (Drummond Scientific, model: Pipet-Aid®, catalog number: 4-000-101 )
- Pipettors (MIDSCI, Alphapette, models: A-20 , A-100 , A-200 , A-1000 )
- 37 °C, 6% CO2 incubator
- 56 °C water bath
- Refrigerator (4 °C)
- -20 °C freezer
- Spectrophotometer compatible with absorbance measurements at 600 nm (Cole-Parmer, Jenway, model: 6320D )
- Beckman Allegra 6KR centrifuge (Beckman Coulter, model: Allegra® 6KR KneewellTM)
- GH-3.8 rotor (Beckman Coulter, model: GH-3.8 Rotor )
- Inverted microscope with enough resolution to detect cells of macrophage size (Olympus, model: IMT-2 with 10x objective)
- Beckman Microfuge® 18 Centrifuge (Beckman Coulter, model: Microfuge® 18 )
- Flow cytometer (BD, model: FACS LSR II )
Software
- FlowJo (BD)
Procedure
- Bacterial culture
Mycobacterium tuberculosis strains are grown at 37 °C in 7H9 OADC media (see Recipes) in standing culture in T25 flasks until mid-log phase (OD600 ~0.6). Bacterial cultures are started from frozen stocks and maintained for no more than 5 passages. Constitutive expression of fluorescent protein by Mtb is required for further detection. We used Mtb Erdman strains with an integrated pMV306 plasmid expressing mCherry from smyc promoter (smyc’::mCherry). This plasmid confers kanamycin resistance, therefore growth media contains kanamycin at a final concentration of 25 μg/ml (see Recipes).
Notes:- Do not grow more than 10 ml of bacterial culture in a T25 flask to ensure sufficient amount of oxygen available to Mtb. To estimate timing for bacterial culture growth, consider that Mtb divides approximately once in two days when grown in standing culture. More details on bacterial culture growth for macrophage infection can be found in (Nazarova and Russell, 2017).
- We advise testing your wild type strain constitutively expressing fluorescent protein for ability to efficiently intake Bodipy-palmitate during macrophage infection by microscopy. We have noticed that in Mtb Erdman strain expression of mCherry under strong promoter (smyc’ or hsp60’) from replicating plasmid impacts Bodipy-palmitate uptake. However CDC1551 strain with the same plasmid demonstrated high level of fatty acid uptake. Additionally, care should be taken to determine by confocal microscopy if Bodipy-palmitate accumulates within bacteria versus on their surface.
- Do not grow more than 10 ml of bacterial culture in a T25 flask to ensure sufficient amount of oxygen available to Mtb. To estimate timing for bacterial culture growth, consider that Mtb divides approximately once in two days when grown in standing culture. More details on bacterial culture growth for macrophage infection can be found in (Nazarova and Russell, 2017).
- Macrophage isolation and culturing
Macrophages of various origins can be used. We differentiated macrophages from bone marrow cells from BALB/c mice and maintained in BMDM media (see Recipes) with antibiotics at 37 °C and 6.0% CO2 for 10 days before infection.
Infection- The day before infection seed macrophages in antibiotic-free BMDM media into T150 tissue culture flasks (3 x 107 cells and 40-50 ml of BMDM media per flask) to obtain confluent monolayers. If macrophages are of limited numbers one T75 flask with 1 x 107 cells would provide sufficient results, however, two T150 flasks (6 x 107 cells) would give easily detectable pellet of bacteria at the end of the experiment.
- On the day of infection measure optical density of mid-log Mtb culture. Assuming that OD600 of 0.6 equals 108 bacteria/ml, centrifuge required amount of culture at 3,300 rpm (~2,500 x g) for 12 min in Beckman Allegra 6KR centrifuge, GH-3.8 rotor. We infect macrophages at MOI of 4:1, therefore to infect 6 x 107 cells we need 2.4 x 108 bacteria. To ensure that sufficient numbers of bacteria are pelleted we routinely centrifuge 3 ml of bacteria at OD600 = 0.6.
Notes: As estimation of bacterial numbers may vary, one may want to determine it on their own. However, to achieve higher replicability in terms of MOI, we advise adhering to our estimation. More details on macrophage infection can be found in (Nazarova and Russell, 2017). - Following centrifugation, remove the supernatant and resuspend the bacterial pellet in 1.5 ml of BUB (see Recipes). Pass bacterial suspension through a 1 ml tuberculin syringe with 25 gauge needle 12-20 times the same syringe and needle. Add 3.5 ml of BUB to the suspension to obtain 5 ml in total and mix thoroughly.
- Add 2.4 ml of bacterial suspension to each T150 flask containing 3 x 107 cells. Mix well, but gently. Incubate at 37 °C and 6% CO2 for 4 h.
- Remove extracellular bacteria by replacing with fresh pre-warmed antibiotic free BMDM media. Infected macrophages are maintained in BMDM medium at 37 °C and 6% CO2 for 3 days. Changing media on the second day of infection is optional.
Labeling- On the day of labeling (third day of infection) pre-warm sterile 1% fatty acid-free BSA (see Recipes) in PBS at 37 °C for 30-60 min. Add 4 mM stock of Bodipy-palmitate (see Recipes) to obtain 100 μM concentration. For labeling one T75 flask add 50 μl of 4 mM Bodipy-palmitate stock to 1.95 ml of 1% fatty acid-free BSA. Mix well by vortexing until the solution turns green. Keep at 37 °C and protect from light.
Notes:- The day of labeling can be chosen with consideration of the questions you are trying to address. We tested uptake of Bodipy-palmitate at various stages of macrophage infection, and noted that before the third day of infection the uptake levels are not sufficient to be detected.
- In addition to Bodipy-palmitate, we have tested Bodipy FL C12 and Bodipy 558/568 C12. Bodipy FL C12 accumulated in intracellular bacteria as efficiently as Bodipy-palmitate, while Bodipy 558/568 C12 was poorly assimilated by Mtb. We chose to use Bodipy-palmitate as fatty acids of the length (16 carbons) are more commonly found in the membranes of macrophages infected with Mtb.
- The day of labeling can be chosen with consideration of the questions you are trying to address. We tested uptake of Bodipy-palmitate at various stages of macrophage infection, and noted that before the third day of infection the uptake levels are not sufficient to be detected.
- For labeling one T75 flask add 1.6 ml of 100 μM Bodipy-palmitate in 1% fatty acid-free BSA (from Step 1 of Labeling) to 18.4 ml of pre-warmed cuvette buffer (see Recipes) such that the final concentration of the labeled lipid is 8 μM, mix well. Use 15 ml for labeling one T75 flask, and 30 ml for labeling one T150 flask. Increase volumes in Steps 1 and 2 (Labeling) accordingly for larger infections.
- Remove media from infected macrophages and replace with cuvette buffer containing Bodipy-palmitate (volumes are described in Step 2 of Labeling). Incubate the infected macrophages with label at 37 °C and 6.0% CO2 for 1 h.
- After the 1 h labeling period, remove the cuvette buffer containing the Bodipy-palmitate and add fresh pre-warmed cuvette buffer without label for 1 h. Use 15 ml for one T75 flask, and 30 ml for one T150 flask. Proceed to the next step right after 1 h incubation without label.
Note: Alternatively, Bodipy-palmitate in 1% fatty acid-free BSA can be added directly to infected macrophages cultured in BMDM media. Label chase can be performed in fresh pre-warmed BMDM medium as well. No cuvette buffer is needed in this case. From our experience, either way of labeling gives comparable results.
Isolation of intracellular bacteria
Note: This portion of the protocol is based on phagosome isolation described in (Pethe et al., 2004).- Remove cuvette buffer from labeled infected cells, and quickly rinse with 10 ml of Homogenization buffer (see Recipes).
- Add 15 ml of ice-cold Homogenization buffer, incubate at 4 °C for 10-15 min, and harvest macrophages by scraping off from each flask with cell scrapers. Transfer the cells to a 50 ml conical tube and pellet the cells by centrifugation at 1,500 rpm (514 x g) for 10 min in Beckman Allegra 6KR centrifuge, GH-3.8 rotor. This and all the following centrifugations are done at 4 °C to block any further uptake of lipids by bacteria.
- Remove supernatant, resuspend pellet in 1.5 ml of Homogenization buffer by pipetting and transfer the cells to a 15 ml conical tube. Lyse the cells by 25-70 passages through a 1 ml tuberculin syringe with 25 gauge needle. Monitor cell lysis under a microscope using glasstic slides with 100 grids. Continue until > 95% of the cells are lysed, when intact cells are replaced by cell debris.
Note: For safety purposes place glass slide with infected material in 150 x 15 mm Petri dish. - Increase the volume to 5 ml by adding Homogenization buffer, resuspend well. Centrifuge cell lysate at 800 rpm (~146 x g) for 10 min in Beckman Allegra 6KR centrifuge, GH-3.8 rotor.
- Transfer the supernatant (suspension of phagosomes) into a new 15 ml conical tube. The pellet mainly consists of nuclei and unlysed cells and is discarded.
- To the suspension add 20% Tween 80 (see Recipes) to a final concentration of 0.1%, mix well and leave at 4 °C for 15 min to lyse Mtb containing vacuoles.
- Quickly agitate by shaking and isolate the bacteria by centrifugation at 2,500 rpm (1,430 x g) for 15 min in Beckman Allegra 6KR centrifuge, GH-3.8 rotor.
- Remove supernatant and resuspend the bacterial pellet in 10 ml of 0.05% tyloxapol in PBS (see Recipes). Centrifuge bacteria down at 3,300 rpm (~2,500 x g) for 15 min in Beckman Allegra 6KR centrifuge, GH-3.8 rotor.
- Optional: repeat the previous step to further remove labeled fatty acids adhered to bacterial cell surface.
Note: Non-specific binding of bodipy-palmitate to the bacterial cell surface produces background signal. However, if tested strains/conditions produce a significant difference in specific uptake of label, this background noise wouldn’t impact results greatly. - Remove supernatant and fix bacteria in 4% PFA (see Recipes) for 24 h in 2 ml screw-cap tube.
Note: If you have flow cytometer in BSL3 facility, bacteria can be analyzed live immediately after isolation and without fixation.
- The day before infection seed macrophages in antibiotic-free BMDM media into T150 tissue culture flasks (3 x 107 cells and 40-50 ml of BMDM media per flask) to obtain confluent monolayers. If macrophages are of limited numbers one T75 flask with 1 x 107 cells would provide sufficient results, however, two T150 flasks (6 x 107 cells) would give easily detectable pellet of bacteria at the end of the experiment.
Data analysis
Isolated bacteria should be analyzed within few days following collection, ideally the next day.
- Centrifuge fixed samples at 10,000 rpm (9,000 x g) for 5 min in Beckman Microfuge® 18 Centrifuge.
- Remove supernatant, resuspend pellet in 1-2 ml of 0.05% tyloxapol in PBS and transfer into FACS tube.
- Pass suspension through 1 ml tuberculin syringe with 25 gauge needle 12-20 times to obtain a single cell bacterial suspension.
- Analyze immediately on flow cytometer (BD FACS LSR II ) using described gating strategy (Figure 2). Select population of a medium size in forward and side scatter to exclude clumps and small debris, focus on mCherry(PE-Texas Red)-positive population (bacteria), and compare FITC signal from the Bodipy-palmitate between your samples. Since there is minimal overlap between mCherry and Bodipy signals, compensation is not needed. As a negative control use mCherry-positive bacteria not exposed to labeling.
Notes:- Bodipy-palmitate also accumulates in membranes of cell organelles, which are excluded from analysis, because they are mCherry-negative.
- Collect as many events as possible, minimum 50,000. For analysis in Figure 2 we collected 1,000,000 events.
- Bodipy-palmitate also accumulates in membranes of cell organelles, which are excluded from analysis, because they are mCherry-negative.
- Quantify acquired data using FlowJo by determining mean fluorescence of Bodipy signal from mCherry-positive bacteria.
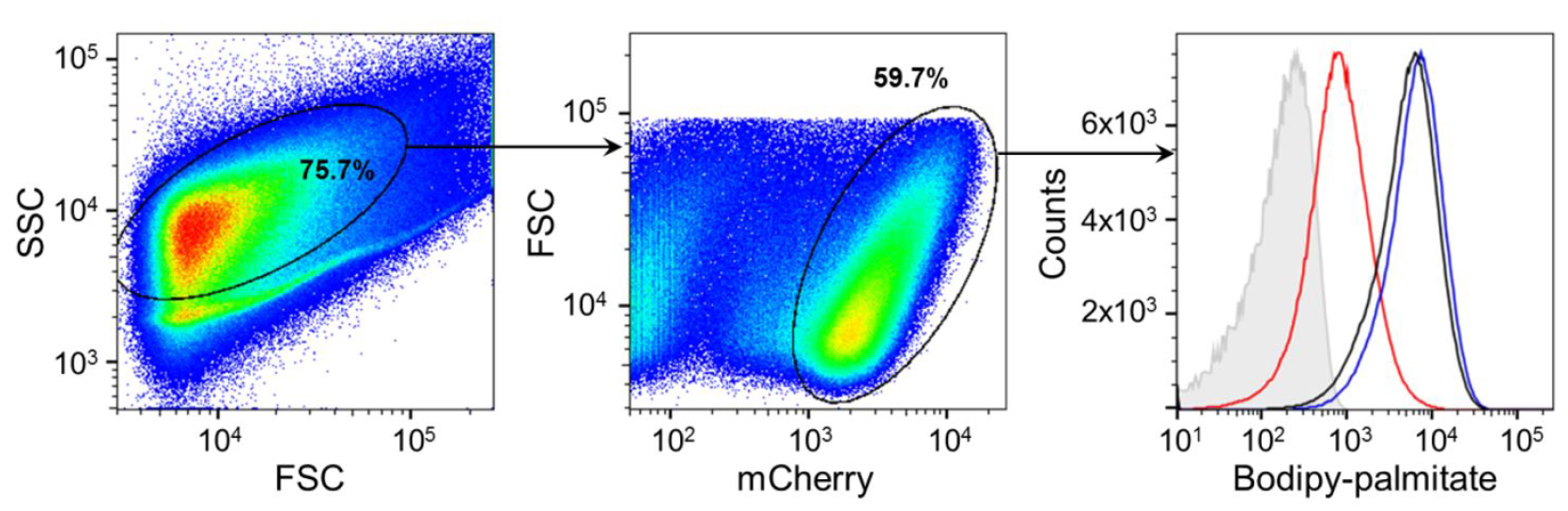
Figure 2. Gating strategy for analysis of Bodipy-palmitate uptake by intracellular Mtb. The population of a medium size is selected in forward and side scatter, and analyzed further for level of mCherry signal. Bodipy palmitate signal is determined for the mCherry-positive population representing bacteria. Panel on the right is a representative of detected Bodipy-palmitate signal associated with three different strains: wild type (black), ∆lucA::hyg (red) and complemented strain (blue). Grey histogram represents mCherry-positive bacteria not exposed to labeling. (Adapted from Nazarova et al., 2017)
Recipes
- 20% tyloxapol (20 ml)
- Using 3 ml syringe add 4 ml of tyloxapol to 16 ml of distilled water in 50 ml conical tube
- Heat up at 56 °C, vortex occasionally, until tyloxapol goes into a viscous but clear solution
- Filter-sterilize (0.22 μm), store at room temperature for up to 12 months
- Using 3 ml syringe add 4 ml of tyloxapol to 16 ml of distilled water in 50 ml conical tube
- 7H9 OADC media (1 L)
- Dissolve 4.7 g 7H9 DifcoTM Middlebrook 7H9 Broth Base and 2 ml glycerol in 900 ml of distilled water
- Aseptically add 2.5 ml of sterile 20% tyloxapol to a final concentration of 0.05% and 100 ml of BBLTM Middlebrook OADC Enrichment, mix
- Filter-sterilize (0.22 μm), store at room temperature
- Dissolve 4.7 g 7H9 DifcoTM Middlebrook 7H9 Broth Base and 2 ml glycerol in 900 ml of distilled water
- Kanamycin 25 mg/ml (10 ml)
- Add 250 mg kanamycin sulfate to 10 ml distilled water, mix by vortexing
- Filter-sterilize (0.22 μm), aliquot and store at -20 °C
- Add 250 mg kanamycin sulfate to 10 ml distilled water, mix by vortexing
- D10 media (1 L)
- Add 100 ml heat inactivated fetal bovine serum (10% final concentration), 10 ml 200 mM L-glutamine (2 mM final), 10 ml 100 mM sodium pyruvate (1 mM final), 10 ml penicillin, 100x streptomycin solution to Dulbecco’s modified Eagle’s medium DMEM so that total volume is 1 L
- Filter-sterilize (0.22 μm), store at 4 °C for no longer than 2 months
- Preheat at 37 °C before use
- Add 100 ml heat inactivated fetal bovine serum (10% final concentration), 10 ml 200 mM L-glutamine (2 mM final), 10 ml 100 mM sodium pyruvate (1 mM final), 10 ml penicillin, 100x streptomycin solution to Dulbecco’s modified Eagle’s medium DMEM so that total volume is 1 L
- L-cell conditioned media
- Frozen cells are thawed into D10 media and grown for 12-14 days in T150 tissue culture flasks at 37 °C and 6% CO2
- Conditioned media is collected, and cell debris is removed by centrifugation at 1,500 rpm (514 x g) for 10 min on Beckman Allegra 6KR centrifuge, GH-3.8 rotor
- Supernatant is aliquoted and stored at -20 °C
- Frozen cells are thawed into D10 media and grown for 12-14 days in T150 tissue culture flasks at 37 °C and 6% CO2
- BMDM media (1 L)
- Add 100 ml heat inactivated fetal bovine serum (10% final concentration), 10 ml 200 mM L-glutamine (2 mM final), 10 ml 100 mM sodium pyruvate (1 mM final), 100 ml L-cell-conditioned media (10% final concentration) to Dulbecco’s modified Eagle’s medium DMEM so that total volume is 1 L. Add 10 ml penicillin-streptomycin solution (100x) for media to culture macrophages before infection
- Filter-sterilize (0.22 μm), store at 4 °C for no longer than 2 months
- Preheat at 37 °C before use
- Add 100 ml heat inactivated fetal bovine serum (10% final concentration), 10 ml 200 mM L-glutamine (2 mM final), 10 ml 100 mM sodium pyruvate (1 mM final), 100 ml L-cell-conditioned media (10% final concentration) to Dulbecco’s modified Eagle’s medium DMEM so that total volume is 1 L. Add 10 ml penicillin-streptomycin solution (100x) for media to culture macrophages before infection
- Basal uptake buffer (BUB)
- Add 2.25 g glucose, 2.5 g bovine serum albumin, 0.5 ml gelatin, 50 mg CaCl2, 50 mg MgCl2 to 500 ml PBS, mix
- Filter-sterilize (0.22 μm), store at 4 °C for no longer than 6 months
- Add 2.25 g glucose, 2.5 g bovine serum albumin, 0.5 ml gelatin, 50 mg CaCl2, 50 mg MgCl2 to 500 ml PBS, mix
- 1% fatty acid-free BSA (50 ml)
- Dissolve 500 mg of fatty acid-free BSA in 50 ml PBS
- Filter-sterilize (0.22 μm), aliquot and store at -20 °C
- Dissolve 500 mg of fatty acid-free BSA in 50 ml PBS
- 4 mM Bodipy-palmitate
- Add 526 μl of 100% ethanol to 1 mg of Bodipy-palmitate, mix by vortexing
- Store at -20 °C, protect from light
- Add 526 μl of 100% ethanol to 1 mg of Bodipy-palmitate, mix by vortexing
- Cuvette buffer (1 L)
- Dissolve 101 mg CaCl2, 200 mg KCl, 102 mg MgCl2, 901 mg glucose in 1 L of PBS
- Filter-sterilize (0.22 μm) and store at room temperature
- A few days before use add heat inactivated fetal bovine serum to final concentration of 10% to obtain the desired volume, filter-sterilize (0.22 μm) and store at 4 °C
- Preheat at 37 °C before use
- Dissolve 101 mg CaCl2, 200 mg KCl, 102 mg MgCl2, 901 mg glucose in 1 L of PBS
- Homogenization buffer (500 ml)
- Dissolve 42.75 g sucrose, 95 mg EGTA, 2.38 g HEPES, 250 μl gelatin in 400 ml of distilled water
- Adjust pH to 7.0
- Fill up with distilled water to the final volume 500 ml
- Filter-sterilize (0.22 μm). Store and keep throughout use at 4 °C
- Dissolve 42.75 g sucrose, 95 mg EGTA, 2.38 g HEPES, 250 μl gelatin in 400 ml of distilled water
- 20% Tween 80 (20 ml)
- Using 3 ml syringe add 4 ml of Tween 80 to 16 ml of distilled water in 50 ml conical tube
- Heat up at 37 °C, vortex occasionally, until Tween 80 goes into solution
- Filter-sterilize (0.22 μm), store at room temperature
- Using 3 ml syringe add 4 ml of Tween 80 to 16 ml of distilled water in 50 ml conical tube
- 0.05% tyloxapol (250 ml)
- Aseptically add 625 μl of sterile 20% tyloxapol to 250 ml PBS, mix
- Filter-sterilize (0.22 μm), store at room temperature
- Aseptically add 625 μl of sterile 20% tyloxapol to 250 ml PBS, mix
- 4% PFA (500 ml)
- Mix 20 g of PFA in PBS while heating up. Avoid boiling, otherwise formaldehyde will be formed
- Aliquot and store at -20 °C. Thaw at room temperature before use
- Mix 20 g of PFA in PBS while heating up. Avoid boiling, otherwise formaldehyde will be formed
Acknowledgments
We thank Linda Bennett for excellent technical support. This work was supported by the NIH grants (AI099569 and AI119122) to BCV and (AI080651 and AI134183) to DGR. This protocol was developed and reported in the previous publication (Nazarova et al., 2017). The authors declare no conflicts of interest or competing interests.
References
- Caire-Brändli, I., Papadopoulos, A., Malaga, W., Marais, D., Canaan, S., Thilo, L. and de Chastellier, C. (2014). Reversible lipid accumulation and associated division arrest of Mycobacterium avium in lipoprotein-induced foamy macrophages may resemble key events during latency and reactivation of tuberculosis. Infect Immun 82(2): 476-490.
- Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., Gordon, S. V., Eiglmeier, K., Gas, S., Barry, C. E., 3rd, Tekaia, F., Badcock, K., Basham, D., Brown, D., Chillingworth, T., Connor, R., Davies, R., Devlin, K., Feltwell, T., Gentles, S., Hamlin, N., Holroyd, S., Hornsby, T., Jagels, K., Krogh, A., McLean, J., Moule, S., Murphy, L., Oliver, K., Osborne, J., Quail, M. A., Rajandream, M. A., Rogers, J., Rutter, S., Seeger, K., Skelton, J., Squares, R., Squares, S., Sulston, J. E., Taylor, K., Whitehead, S. and Barrell, B. G. (1998). Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393(6685): 537-544.
- Daniel, J., Maamar, H., Deb, C., Sirakova, T. D. and Kolattukudy, P. E. (2011). Mycobacterium tuberculosis uses host triacylglycerol to accumulate lipid droplets and acquires a dormancy-like phenotype in lipid-loaded macrophages. PLoS Pathog 7(6): e1002093.
- Fontán, P., Aris, V., Ghanny, S., Soteropoulos, P. and Smith, I. (2008). Global transcriptional profile of Mycobacterium tuberculosis during THP-1 human macrophage infection. Infect Immun 76(2): 717-725.
- Forrellad, M. A., McNeil, M., Santangelo Mde, L., Blanco, F. C., Garcia, E., Klepp, L. I., Huff, J., Niederweis, M., Jackson, M. and Bigi, F. (2014). Role of the Mce1 transporter in the lipid homeostasis of Mycobacterium tuberculosis. Tuberculosis (Edinb) 94(2): 170-177.
- Homolka, S., Niemann, S., Russell, D. G. and Rohde, K. H. (2010). Functional genetic diversity among Mycobacterium tuberculosis complex clinical isolates: delineation of conserved core and lineage-specific transcriptomes during intracellular survival. PLoS Pathog 6(7): e1000988.
- Lovewell, R. R., Sassetti, C. M. and VanderVen, B. C. (2016). Chewing the fat: lipid metabolism and homeostasis during M. tuberculosis infection. Curr Opin Microbiol 29: 30-36.
- Nazarova, E. V., Montague, C. R., La, T., Wilburn, K. M., Sukumar, N., Lee, W., Caldwell, S., Russell, D. G. and VanderVen, B. C. (2017). Rv3723/LucA coordinates fatty acid and cholesterol uptake in Mycobacterium tuberculosis. Elife 6.
- Nazarova, E. V. and Russell, D. G. (2017). Growing and handling of Mycobacterium tuberculosis for macrophage infection assays. Methods Mol Biol 1519: 325-331.
- Pandey, A. K. and Sassetti, C. M. (2008). Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci 105: 4376-4380.
- Pethe, K., Swenson, D. L., Alonso, S., Anderson, J., Wang, C. and Russell, D. G. (2004). Isolation of Mycobacterium tuberculosis mutants defective in the arrest of phagosome maturation. Proc Natl Acad Sci U S A 101(37): 13642-13647.
- Podinovskaia, M., Lee, W., Caldwell, S. and Russell, D. G. (2013). Infection of macrophages with Mycobacterium tuberculosis induces global modifications to phagosomal function. Cell Microbiol 15(6): 843-859.
- Rachman, H., Strong, M., Ulrichs, T., Grode, L., Schuchhardt, J., Mollenkopf, H., Kosmiadi, G. A., Eisenberg, D. and Kaufmann, S. H. (2006). Unique transcriptome signature of Mycobacterium tuberculosis in pulmonary tuberculosis. Infect Immun 74(2): 1233-1242.
- Rohde, K. H., Abramovitch, R. B. and Russell, D. G. (2007). Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe 2(5): 352-364.
- Rohde, K. H., Veiga, D. F., Caldwell, S., Balazsi, G. and Russell, D. G. (2012). Linking the transcriptional profiles and the physiological states of Mycobacterium tuberculosis during an extended intracellular infection. PLoS Pathog 8(6): e1002769.
- Russell, D. G., VanderVen, B. C., Lee, W., Abramovitch, R. B., Kim, M. J., Homolka, S., Niemann, S. and Rohde, K. H. (2010). Mycobacterium tuberculosis wears what it eats. Cell Host Microbe 8(1): 68-76.
- Schnappinger, D., Ehrt, S., Voskuil, M. I., Liu, Y., Mangan, J. A., Monahan, I. M., Dolganov, G., Efron, B., Butcher, P. D., Nathan, C. and Schoolnik, G. K. (2003). Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: Insights into the phagosomal environment. J Exp Med 198(5): 693-704.
- Tailleux, L., Waddell, S. J., Pelizzola, M., Mortellaro, A., Withers, M., Tanne, A., Castagnoli, P. R., Gicquel, B., Stoker, N. G., Butcher, P. D., Foti, M. and Neyrolles, O. (2008). Probing host pathogen cross-talk by transcriptional profiling of both Mycobacterium tuberculosis and infected human dendritic cells and macrophages. PLoS One 3(1): e1403.
- VanderVen, B. C., Fahey, R. J., Lee, W., Liu, Y., Abramovitch, R. B., Memmott, C., Crowe, A. M., Eltis, L. D., Perola, E., Deininger, D. D., Wang, T., Locher, C. P. and Russell, D. G. (2015). Novel inhibitors of cholesterol degradation in Mycobacterium tuberculosis reveal how the bacterium's metabolism is constrained by the intracellular environment. PLoS Pathog 11(2): e1004679.
- Wipperman, M. F., Sampson, N. S. and Thomas, S. T. (2014). Pathogen roid rage: cholesterol utilization by Mycobacterium tuberculosis. Crit Rev Biochem Mol Biol 49(4): 269-293.
Article Information
Copyright
Nazarova et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Nazarova, E. V., Podinovskaia, M., Russell, D. G. and VanderVen, B. C. (2018). Flow Cytometric Quantification of Fatty Acid Uptake by Mycobacterium tuberculosis in Macrophages. Bio-protocol 8(4): e2734. DOI: 10.21769/BioProtoc.2734.
- Nazarova, E. V., Montague, C. R., La, T., Wilburn, K. M., Sukumar, N., Lee, W., Caldwell, S., Russell, D. G. and VanderVen, B. C. (2017). Rv3723/LucA coordinates fatty acid and cholesterol uptake in Mycobacterium tuberculosis. Elife 6.
Category
Microbiology > Microbial metabolism > Lipid
Immunology > Immune cell function > Macrophage
Cell Biology > Cell metabolism > Lipid
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link


.jpg)







