- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Determination of DNA Damage in the Retina Photoreceptors of Drosophila
Published: Vol 8, Iss 3, Feb 5, 2018 DOI: 10.21769/BioProtoc.2708 Views: 7113
Reviewed by: Oneil G. BhalalaSandhya GanesanAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Intravitreal NHS-Biotin Injection and Immunohistochemistry to Label and Image Protein Transport in the Mouse Optic Nerve
Caroline R. McKeown [...] Hollis T. Cline
Aug 20, 2025 2696 Views

Colocalizing Telomeres With PML or γH2AX Foci by IF-FISH in Mouse Brain Neurons
Anna Konopka
Nov 5, 2025 1506 Views

A Quantitative DNA Fiber Assay to Monitor Replication Fork Progression, Protection, and Restart
Debanjali Bhattacharya and Ganesh Nagaraju
Feb 5, 2026 146 Views
Abstract
The retina is sensitive for light damages, because of direct light exposure, especially intense blue and UV light, which increase level of ROS and other toxic phototransduction products in photoreceptor cells. In our previous work (Damulewicz et al., 2017a and 2017b), we used 8-oxo-deoxyguanosine (8-OHdG) as a marker for oxidative stress to investigate the role of heme oxygenase in DNA protection against UV light. In this protocol, we showed how to determine the level of DNA damages in the retina using immunohistochemical staining.
Keywords: Oxidative stressBackground
Deoxyribonucleic acid (DNA) is an essential molecule for all living organisms. It contains genetic code with instruction for proteins and other molecules necessary for structure and metabolism of an organism. DNA is composed of two strands of nucleotides. Each nucleotide is built of one of four types of nucleobases (cytosine, guanine, adenine and thymine), deoxyribose and phosphate group. Physical and chemical changes in DNA structure are classified as DNA damages. It includes one or two strands of DNA breaks, missing nucleotide or chemical changes of nucleobases. Damages can be induced by environmental factors, like chemicals or UV light exposure, or in result of metabolic processes, which produce reactive oxygen species, reactive nitrogen species, reactive carbonyl species and alkylating agents (De Bont and van Larebeke, 2004).
One of the most common DNA damage is 8-oxo-deoxyguanosine (8-OHdG), which is a product of DNA oxidation by reactive oxygen species and it serves as a marker of oxidative stress (De Souza-Pinto et al., 2001; Swenberg et al., 2011; Valavanidis et al., 2013). 8-OHdG enhances the risk of transversion mutation, because it has ability to pair with adenine and cytosine bases (Maki and Sekiguchi, 1992). In effect it plays a role in mutagenesis, cancerogenesis and aging (De Bont and van Larebeke, 2004; Kohen and Nyska, 2002). Higher 8-OHdG level was described in breast, renal and gastric tumors (Lee et al., 1998; Musarrat et al., 1996; Okamoto et al., 1994).
There are several methods to determine 8-OHdG level in a specific tissue, like immunohistochemistry (De Carvalho et al., 2013), Western blot (Kannan et al., 2006), standard HPLC (Herrero and Barja, 1999), liquid chromatography nanoelectrospray-tandem mass spectrometry (Ma et al., 2016). Here, we present a detailed protocol for DNA damage determination in the retina of fruit fly, Drosophila melanogaster using immunohistochemistry method (Damulewicz et al., 2017a and 2017b).
Materials and Reagents
- Filter paper (Fisher Scientific, catalog number: 09-790-14D )
- Cover glasses (Menzel-Glaser)
- Microscope glasses (Superfrost, Menzel-Glaser)
- Small plastic holders (i.e., Eppendorf tube lids)
- Tissue-Tek (SAKURA, catalog number: 4583 )
- Drosophila melanogaster (wild-type CantonS, 5-7 days old males)
Note: Local ethical standards need to be followed.
- 6% glucose in water (commercial)
- Protoporphyrin IX (SnPP IX) (Frontier Scientific, catalog number: P562-9 )
- Hemin chloride (Merck, Calbiochem, catalog number: 3741 )
- CO2
- Schneider’s medium (Sigma-Aldrich, catalog number: S9895 )
- Etoposide (Sigma-Aldrich, catalog number: E1383 )
- 4% paraformaldehyde (PFA) (Sigma-Aldrich, catalog number: P6148 )
- Sucrose (commercial)
- Liquid nitrogen
- Normal Goat Serum (NGS) (Sigma-Aldrich, catalog number: G9023 )
- Bovine serum albumin (BSA)
- Mouse anti-8-hydroxyguanosine primary antibodies (Acris, catalog number: AM03160 )
- HRP/DAB (ABC) kit (Abcam, catalog number: ab64264 )
- Glycerol (MP Biomedicals)
- Vectashield with DAPI (Vector Laboratories, catalog number: H-1200 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S7653 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9541 )
- Sodium phosphate monobasic monohydrate (NaH2PO4·H2O) (Sigma-Aldrich, catalog number: S3522 )
- Potassium phosphate monobasic (KH2PO4) (Sigma-Aldrich, catalog number: P5655 )
- Hydrochloric acid (HCl) (Sigma-Aldrich, catalog number: H1758 )
- Sodium phosphate dibasic (Na2HPO4) (Sigma-Aldrich, catalog number: S7907 )
- Triton X-100 (Sigma-Aldrich, catalog number: T8787 )
- Cornmeal (Glutenex)
- Agar (Bioshop)
- Artificial honey (Vortumnus)
- Yeast (Paneo)
- Methyl-4-hydroxybenzoate (anti-fungal chemical, Sigma-Aldrich, catalog number: H3647 )
- 90% EtOH
- Phosphate buffered saline (PBS) (see Recipes)
- Phosphate buffer with Triton (PBT) (see Recipes)
- Standard medium for flies (see Recipes)
Equipment
- Forceps (Dumont No. 5)
- UV transilluminator (Vilbert Lourmat, model: UVECX26 )
- White light transilluminator (Vilbert Lourmat, model: TFX53WL )
- Cryostat (Leica, model: CM1850 UV )
- Light microscope (Carl Zeiss, model: Axioskop 50 )
- Camera (Canon, model: G10 )
Software
- ImageJ software
Procedure
- Take 30 males for every group (experimental and control).
Notes: You can use different control groups, according to your experiment:- Control for the staining conditions: positive (incubated with etoposide) and negative (without primary antibodies).
- Control for UV light effect–flies unexposed to light (kept in constant darkness).
- Control for feeding effect–flies fed with glucose only.
- Control for the staining conditions: positive (incubated with etoposide) and negative (without primary antibodies).
- Keep flies for 5 days in constant darkness (DD) on a standard medium.
- Place flies for 6 h into an empty vial, with no access to food, but with wet filter paper to provide them water ad libitum. Keep them in DD conditions.
- Apply selected chemicals diluted in 6% glucose to a filter paper for 6 h or 6% glucose only for control groups. In this experiment, we used heme oxygenase inhibitor (100 µM protoporphyrin IX–SnPP IX) and activator (100 µM hemin chloride).
Note: Avoid leaving big droplets of a liquid at the bottom of vial–flies can be trapped in liquid and drown.
- Use CO2 to anesthetize one of the control groups.
- Decapitate one of the control groups and put heads for 4 h in 500 µl Schneider’s medium supplemented with 20 µM etoposide at room temperature.
- Expose experimental flies and second control group (fed with glucose only) to 1 h of UV light pulse (100 lux) and then to 3 h of intensive white light (1,500 lux) (Figure 1).
Note: Be sure that flies have access to water during the experiment (wet filter paper).
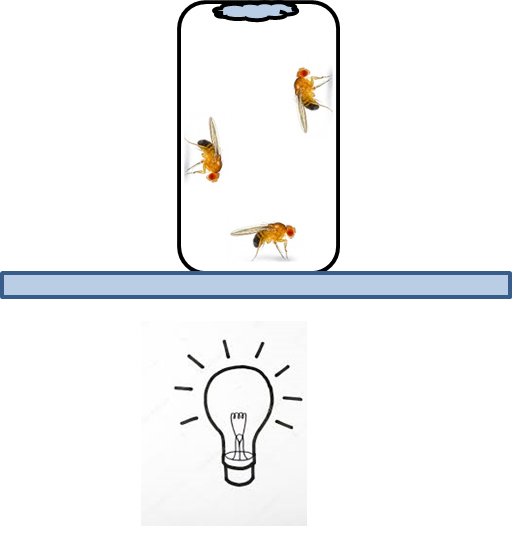
Figure 1. UV/white light exposure. Vial (orientated upside down) with flies is placed on the transilluminator. Filter paper attached to the vial provide water ad libitum.
- Keep one of control group continuously in constant darkness until decapitation.
- Use CO2 to anesthetize flies.
- Decapitate the anesthetized flies (all experimental and control) in a drop of 4% PFA.
- Fix all heads in 500 µl 4% PFA for 4 h at 4 °C.
- Remove PFA using a pipette and wash heads 2 times each for 10 min at room temperature (RT) in 500 µl PBS (see Recipes).
- Cryoprotect material by washing for 10 min in 500 µl of 12.5% sucrose and then overnight in 500 µl of 25% sucrose (Figure 2).
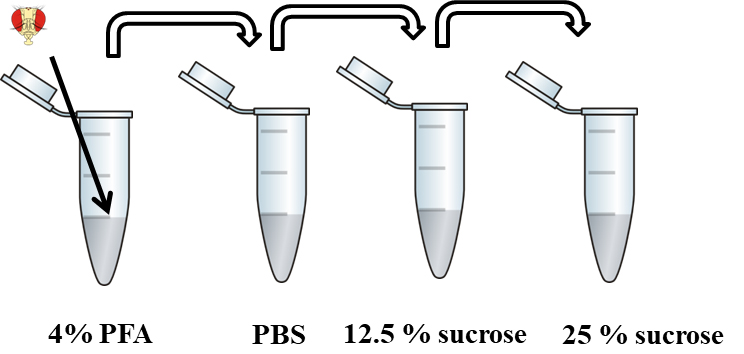
Figure 2. Fixation and cryoprotection of material. Flies heads are fixed in 4% PFA, then washed in PBS and cryoprotected in 12.5% and 25% sucrose.
- Put a droplet of Tissue-Tek to small plastic holders, embed heads into Tissue-Tek, put them into the bottom and organize in rows (Figure 3).
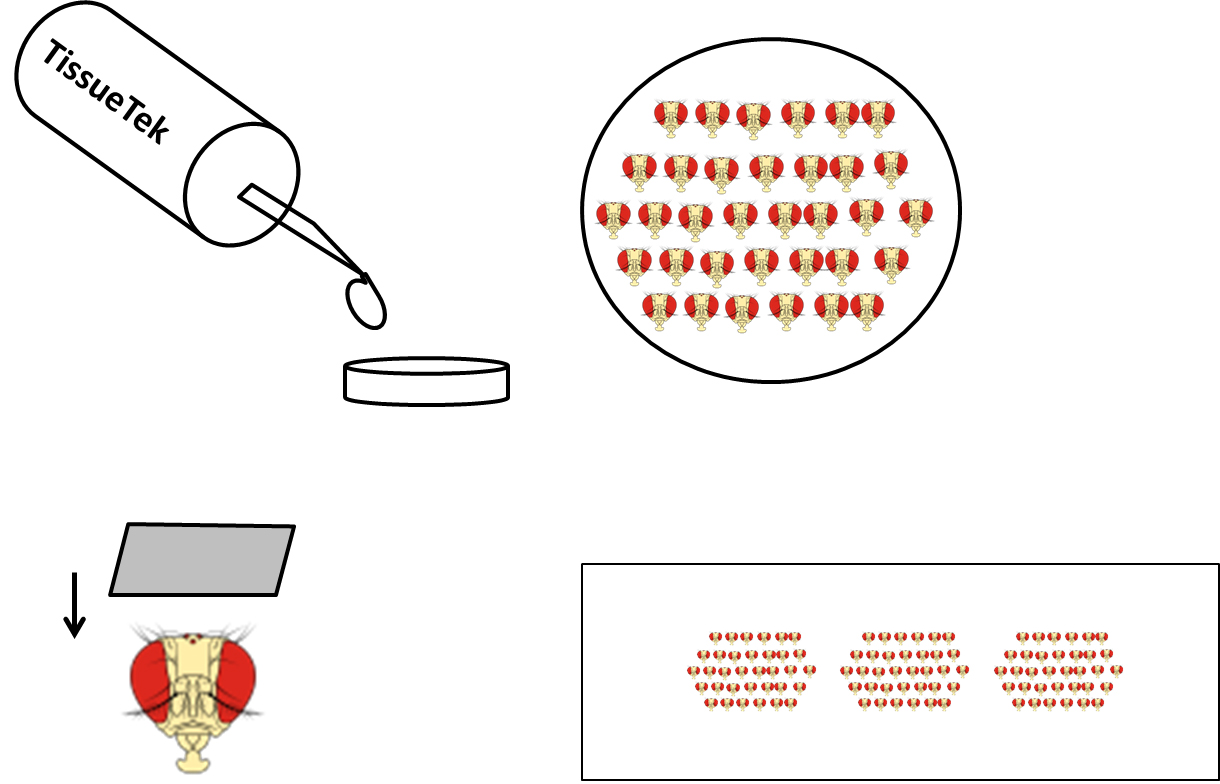
Figure 3. Sample preparation for cryostat. Small plastic holder is filled with TissueTek, then heads are put into the bottom in rows and cut (frontal sections). Sections are collected on microscope glass.
- Freeze material in liquid nitrogen for few seconds.
- Remove frozen sample from the holder.
- Use a cryostat to cut 20 µm thick sections.
- Dry the cryosections for 30 min at room temperature.
- Keep the sections frozen for long-term storage at -20 °C or use them immediately for immunodetection at room temperature.
- Rehydrate material by incubating with 500 µl PBS for 30 min.
Note: Use PBS, PBT at room temperature.
- Wash sections for 10 min in 200 µl 0.5% PBT (see Recipes).
- Wash sections for 5 min in 200 µl 2% PBT.
- Wash sections 3 times for 5 min each in 200 µl 0.5% PBT.
- Block unspecific binding sites by the incubation with 200 µl 5% NGS in BSA for 45 min.
- Apply anti-8-hydroxyguanosine primary antibodies (1:500, in 0.5% BSA) and incubate overnight at 4 °C.
- Keep one section as a negative control and do not apply primary antibodies.
- Use HRP/DAB (ABC) detection kit according to the manufacturer’s protocol. In this step secondary antibodies are used (they are provided in the kit).
- Close sections with glycerol and cover glasses.
- Take the retina pictures under a light microscope using a microscope camera.
Note: To visualize cell nuclei in the retina keep one section unstained with anti-8OHdG. Rehydrate material and close with fluorescence medium–Vectashield with DAPI. Use a fluorescent or confocal microscope to take pictures.
Data analysis
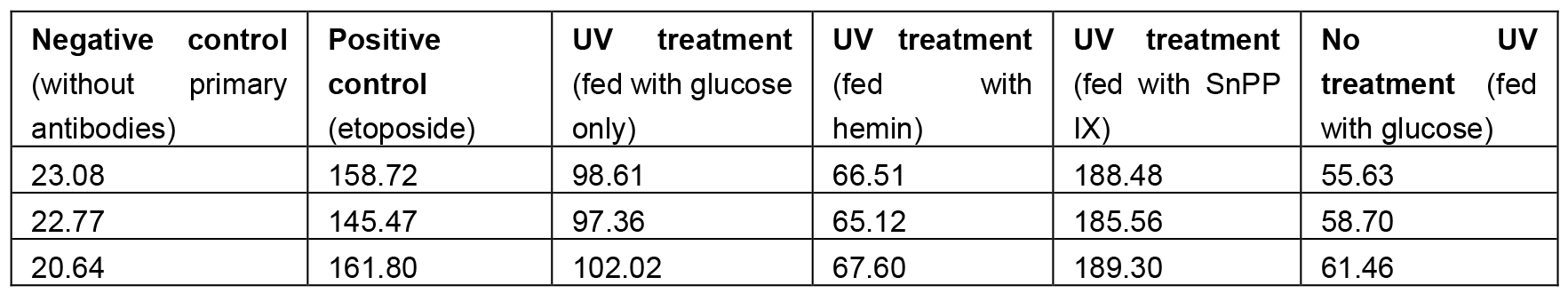
- Analyze DNA damages as Mean Grey Values using ImageJ software. Select area on the top of retina (where nuclei are located, see Figure 5) and measure Mean Grey Value (the range between 0-255). Mean Grey Value is the sum of the grey values of all pixels in the area divided by the number of pixels within selection. Collect three measurements from one picture, use the average as a mean for this picture, collect at least 20 means (different individuals) (Table 1). Copy your data to Excel file and compare means between experimental and control groups.
Note: The retina photoreceptors nuclei are located in the distal part of the retina (see Figures 4 and 5, nuclei are stained with DAPI), measure Mean Grey Value in this area only. Use the same microscope and camera settings for all groups (experimental and control).
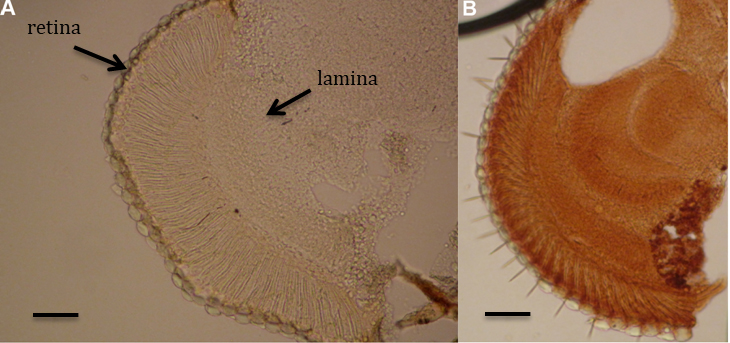
Figure 4. Controls used for DNA damage determination. A. Negative control–without primary antibodies. B. Positive control–etoposide treatment. Scale bars = 20 µm.
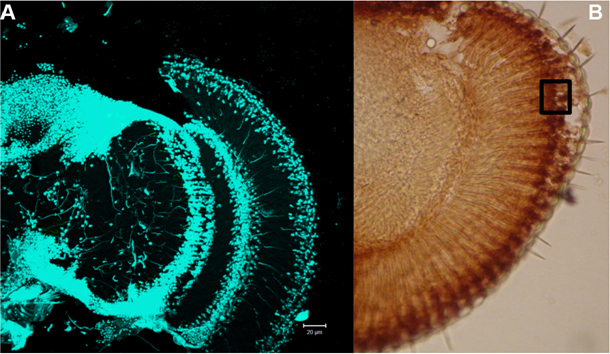
Figure 5. Localization of cell nuclei in the retina. A. DAPI staining–cell nuclei are visualized. B. Retina immunostained with anti-8-hydroxyguanosine, black square shows an analyzed area. Scale bar = 20 µm.
Table 1. Sample triplicate measurements for experimental and control flies
Recipes
- Phosphate buffer (PBS, 1x)
8 g NaCl
0.2 g KCl
1.44 g Na2HPO4
0.24 g KH2PO4
800 ml distilled H2O
Adjust pH to 7.4 with HCl
Adjust volume to 1 L with distilled H2O
- Phosphate buffer with Triton (PBT, 1 L)
0.4363 g NaH2PO4·H2O
0.9705 g Na2HPO4
7.953 g NaCl
5 ml Triton X-100 for 0.5% PBT
Distilled water up to 1 L
Note: PBS and PBT can be stored for a few weeks in the fridge.
- Standard medium for flies
120 g cornmeal (Glutenex)
10 g agar (Bioshop)
140 ml artificial honey (Vortumnus)
10 g yeast (Paneo)
2 L of water
Boil 30 min
Add 3 g methyl-4-hydroxybenzoate (anti-fungal chemical, Sigma-Aldrich) in 15 ml 90% EtOH
Acknowledgments
The study was supported by the Polish National Science Centre (NCN) grant No. UMO-2012/07/B/NZ3/02908 to EP. The protocol was adapted according to our previous papers: Damulewicz, M., Loboda, A., Jozkowicz, A., Dulak, J. and Pyza, E. (2017). Interactions between the circadian clock and heme oxygenase in the retina of Drosophila melanogaster. Mol. Neurobiol 54(7): 4593-4962. Damulewicz, M., Loboda, A., Jozkowicz, A., Dulak, J. and Pyza, E. (2017). Haeme oxygenase protects against UV light DNA damages in the retina in clock-dependent manner. Sci Rep 7(1): 5197. The authors declare that they have no conflicts or competing interests.
References
- Damulewicz, M., Loboda, A., Jozkowicz, A., Dulak, J. and Pyza, E. (2017a). Interactions between the circadian clock and heme oxygenase in the retina of Drosophila melanogaster. Mol Neurobiol 54(7): 4593-4962.
- Damulewicz, M., Loboda, A., Jozkowicz, A., Dulak, J. and Pyza, E. (2017b). Haeme oxygenase protects against UV light DNA damages in the retina in clock-dependent manner. Sci Rep 7(1): 5197.
- De Bont, R. and van Larebeke, N. (2004). Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis 19(3): 169-185.
- de Carvalho, L. F., Abrao, M. S., Biscotti, C., Sharma, R., Agarwal, A. and Falcone, T. (2013). Mapping histological levels of 8-hydroxy-2'-deoxyguanosine in female reproductive organs. J Mol Histol 44(1): 111-116.
- de Souza-Pinto, N. C., Eide, L., Hogue, B. A., Thybo, T., Stevnsner, T., Seeberg, E., Klungland, A. and Bohr, V. A. (2001). Repair of 8-oxodeoxyguanosine lesions in mitochondrial dna depends on the oxoguanine dna glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial dna of OGG1-defective mice. Cancer Res 61(14): 5378-5381.
- Herrero, A. and Barja, G. (1999). 8-oxo-deoxyguanosine levels in heart and brain mitochondrial and nuclear DNA of two mammals and three birds in relation to their different rates of aging. Aging (Milano) 11(5): 294-300.
- Kannan, S., Pang, H., Foster, D. C., Rao, Z. and Wu, M. (2006). Human 8-oxoguanine DNA glycosylase increases resistance to hyperoxic cytotoxicity in lung epithelial cells and involvement with altered MAPK activity. Cell Death Differ 13(2): 311-323.
- Kohen, R. and Nyska, A. (2002). Oxidation of biological systems: oxidative stress phenomena, antioxidants, redox reactions, and methods for their quantification. Toxicol Pathol 30(6): 620-650.
- Lee, B. M., Jang, J. J. and Kim, H. S. (1998). Benzo[a]pyrene diol-epoxide-I-DNA and oxidative DNA adducts associated with gastric adenocarcinoma. Cancer Lett 125(1-2): 61-68.
- Ma, B., Jing, M., Villalta, P. W., Kapphahn, R. J., Montezuma, S. R., Ferrington, D. A. and Stepanov, I. (2016). Simultaneous determination of 8-oxo-2'-deoxyguanosine and 8-oxo-2'-deoxyadenosine in human retinal DNA by liquid chromatography nanoelectrospray-tandem mass spectrometry. Sci Rep 6: 22375.
- Maki, H. and Sekiguchi, M. (1992). MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature 355(6357): 273-275.
- Musarrat, J., Arezina-Wilson, J. and Wani, A. A. (1996). Prognostic and aetiological relevance of 8-hydroxyguanosine in human breast carcinogenesis. Eur J Cancer 32A(7): 1209-1214.
- Okamoto, K., Toyokuni, S., Uchida, K., Ogawa, O., Takenewa, J., Kakehi, Y., et al. (1994). Formation of 8-hydroxy-2’-deoxyguanosine and 4-hydroxy-2-nonenal-modified proteins in human renal-cell carcinoma. Int J Cancer 58: 825-829.
- Swenberg, J. A., Lu, K., Moeller, B. C., Gao, L., Upton, P. B., Nakamura, J. and Starr, T. B. (2011). Endogenous versus exogenous DNA adducts: their role in carcinogenesis, epidemiology, and risk assessment. Toxicol Sci 120 Suppl 1: S130-145.
- Valavanidis, A., Vlachogianni, T., Fiotakis, K. and Loridas, S. (2013). Pulmonary oxidative stress, inflammation and cancer: respirable particulate matter, fibrous dusts and ozone as major causes of lung carcinogenesis through reactive oxygen species mechanisms. Int J Environ Res Public Health 10(9): 3886-3907.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Damulewicz, M. and Pyza, E. (2018). Determination of DNA Damage in the Retina Photoreceptors of Drosophila. Bio-protocol 8(3): e2708. DOI: 10.21769/BioProtoc.2708.
Category
Molecular Biology > DNA > DNA damage and repair
Neuroscience > Neuroanatomy and circuitry > Optic nerve
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link