- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Monitoring the Targeting of Cathepsin D to the Lysosome by Metabolic Labeling and Pulse-chase Analysis
Published: Vol 7, Iss 21, Nov 5, 2017 DOI: 10.21769/BioProtoc.2598 Views: 8071
Reviewed by: Ralph BottcherKate HannanYong Teng

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Preparation of Testicular Cells for Immunofluorescence Analysis of Manchette in Elongating Spermatids
Changmin Niu [...] Zhibing Zhang
Jun 20, 2025 2374 Views

Western Blotting and Immunoprecipitation of Native Human PIEZO1 Channels
Jinyuan Vero Li [...] Charles D. Cox
Jul 20, 2025 3560 Views

Cluster FLISA—A Method to Compare Protein Expression Efficiency Between Cell Lines and Subunit Clustering of Proteins
Sabrina Brockmöller and Lara Maria Molitor
Nov 5, 2025 1207 Views
Abstract
Mannose 6-phosphate receptors function can be studied in living cells by investigating alterations in processing and secretion of their ligand Cathepsin D. The assay described here is well established in the literature and comprises the metabolic labeling of newly synthesized proteins with [35S] methionine-cysteine in HeLa cells to monitor Cathepsin D processing through secretory pathway and secretion using immunoprecipitation, SDS-PAGE and fluorography.
Keywords: Acid hydrolasesBackground
Cathepsin D (catD) is a lysosomal aspartic protease sorted by Mannose 6-phosphate receptors (M6PRs) that transport it from the trans-Golgi network to endosomes/lysosomes in mammalian cells (Ghosh et al., 2003). CatD is synthesized as a precursor protein (~52 kDa), which is cleaved in lysosomes to generate an intermediate (~48 kDa) or the mature lysosomal form (~34 kDa). Trace amounts of the precursor protein are also secreted from the biosynthetic pathway (Benes et al., 2008). The abundance of catD can be determined using several approaches, such as immunofluorescence-based staining (Poole et al., 1972), Western blotting, fluorometric activity assay (Bewley et al., 2011) or metabolic labeling with pulse-chase analysis (Hirst et al., 2009; Kametaka et al., 2007; Tavares et al., 2017). The later is considered a highly sensitive and quantitative approach to monitor catD dynamics (post-translational processing, secretion and degradation) through the secretory pathway. Specifically it involves metabolically labeling newly synthesized proteins in cells followed by a chase, and then to immunoprecipitate catD from the cell lysate and media (secreted form). Therefore, this method provides a way to follow catD molecules from synthesis to lysosomal targeting or secretion with minimal disturbance of normal cell physiology in its natural environment. Herein we described metabolic labeling with [35S] methionine-cysteine in HeLa cells to monitor catD processing, secretion and degradation by using immunoprecipitation, SDS-PAGE and fluorography.
Materials and Reagents
- Filter pipette tips
- 6-well plate (Corning, catalog number: 3516 )
- Ice bucket
- Aluminum foil
- 1.5 ml micro-centrifuge tubes (Corning, Axygen®, catalog number: MCT-150-C )
- WhatmanTM 3030-347 Grade 3 MM Chr cellulose chromatography paper sheet (GE Healthcare, catalog number: 3030-347 )
- Minisart filters pore size 0.22 μm (Sartorius, catalog number: 16534-K )
- HeLa (ATCC, catalog number: CCL-2 )
- 10x PBS (Thermo Fisher Scientific, InvitrogenTM, catalog number: AM9624 )
- Penicillin-streptomycin solution (Thermo Fisher Scientific, GibcoTM, catalog number: 15070063 )
- Fetal bovine serum (FBS) (Thermo Fisher Scientific, catalog number: 12657029 )
- Dulbecco’s modified Eagle’s medium-high glucose-without L-methionine, L-cysteine and L-glutamine (Sigma-Aldrich, catalog number: D0422 )
- L-Methionine (Sigma-Aldrich, catalog number: M9625 )
- L-Cysteine (Sigma-Aldrich, catalog number: 168149 )
- L-Glutamine (Thermo Fisher Scientific, GibcoTM, catalog number: 25030081 )
- EasyTagTM EXPRESS 35S protein labeling mix of both 35S-L-methionine and 35S-L-cysteine (Express Protein Label) (PerkinElmer, catalog number: NEG772007MC )
- Trizma® base (Sigma-Aldrich, catalog number: T1503 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S9888 )
- Ethylenediamine tetraacetic acid (EDTA) (Thermo Fisher Scientific, InvitrogenTM, catalog number: 15575020 )
- Triton X-100 (Sigma-Aldrich, catalog number: T8787 )
- Protease inhibitor cocktail (Sigma-Aldrich, catalog number: P8340 )
- Protein A plus Ultralink resin (Thermo Fisher Scientific, catalog number: 53142 )
- 10% (v/v) bovine serum albumin (BSA) (Thermo Fisher Scientific, InvitrogenTM, catalog number: 15561020 )
- Cathepsin D antibody (EMD Millipore, Calbiochem, catalog number: 219361 )
- L-Glutamine (Thermo Fisher Scientific, GibcoTM, catalog number: 25030149 )
- Sodium dodecyl sulfate (SDS) (Sigma-Aldrich, catalog number: L3771 )
- Glycerol (C3H8O3) (Sigma-Aldrich, catalog number: G5516 )
- Bromophenol blue (Sigma-Aldrich, catalog number: B0126 )
- Methanol (CH3OH) (Sigma-Aldrich, catalog number: 322415 )
- Acetic acid (CH3COOH) (Sigma-Aldrich, catalog number: 320099 )
- Amplify fluorographic reagent (GE Healthcare, catalog number: NAMP100 )
- β-Mercaptoethanol (Sigma-Aldrich, catalog number: M3701 )
- Dulbecco’s modified eagle medium (DMEM) (Thermo Fisher Scientific, GibcoTM, catalog number: 12800017 ) (see Recipes)
- 100x L-methionine/L-cysteine solution (see Recipes)
- Pulse medium (see Recipes)
- Chase medium (see Recipes)
- Lysis buffer (see Recipes)
- Wash buffer (see Recipes)
- 2x sample buffer (see Recipes)
- Fixation solution (see Recipes)
- Running buffer (see Recipes)
Equipment
- Pipettes
- Incubator
- Titer plate shaker (Thermo Fisher Scientific, catalog number: 4625Q )
- Centrifuge for 1.5 ml micro-centrifuge tubes (Thermo Fisher Scientific, model: SorvallTM ST 16 , catalog number: 75004380) with rotor for micro-centrifuge tubes (Thermo Fisher Scientific, catalog number: 75003652 )
- Tube rotator capable of end-over-end inversion of the tubes (Phoenix Luferco, catalog number: AP22 or equivalent)
- A chamber to run mini-gels (Bio-Rad Laboratories, model: Mini-PROTEAN Tetra Cell )
- Gel-dryer (Hoefer, model: GD2000 , Slab Gel Dryer) coupled with a vacuum pump
- Exposure cassette for unmounted screen, 20 x 25 cm (GE Healthcare, catalog number: 63-0035-44 )
- Pharos FX plus molecular imager (Bio-Rad Laboratories, catalog number: 1709450 )
- BAS storage phosphor screen (GE Healthcare, catalog number: 28-9564-82 )
- Appropriate receptacle to dispose of solid and liquid contaminated with 35S (according to the local radiation safety guidelines)
Software
- Quantity One 1-D Analysis Software (Bio-Rad Laboratories)
Procedure
- Radioactive pulse-chase
- Plate 5 x 105 HeLa CCL-2 cells/well (6-well plate) in 2 ml of complete DMEM and incubate for 24 h/37 °C/5% CO2 (Figure 1). Use one well for each time-point or condition.
- Tilt plate and aspirate media, wash cells twice with 1 ml pre-warmed (room temperature) PBS.
- Incubate the cells with 700 µl pre-warmed (37 °C) pulse medium (-Cys/-Met; supplemented with 10 µl/ml 100x L-glutamine, see Recipes) for 30 min/37 °C/5% CO2. Remember to check your local radiation safety guidelines. From this step on, the waste has to be collected separately and discarded according to your local safety guidelines.
- Transfer the plate to an ice bucket covered with aluminum foil.
- Remove the medium and add 700 µl/well ice-cold pulse medium (+Cys 35S/+Met 35S).
- Incubate for 15 min/37 °C/5% CO2.
The length of time for pulse depends on the rate of catD biosynthesis, which may vary according to the cell line. For HeLa cells, 15 min/37 °C is adequate. For other cell lines, the user may have to optimize the time of the pulse. - Transfer the plate to an ice bucket covered with aluminum foil.
- Tilt plate and aspirate media, wash cells twice with 1 ml ice-cold PBS.
- Add 800 µl pre-warmed (37 °C) chase medium (see Recipes).
- Incubate the cells for 5 h/37 °C/5% CO2.
- Transfer the plate to an ice bucket covered with aluminum foil.
- Transfer the culture media (hereafter called ‘Media’ sample) to micro-centrifuge tubes and place them on ice.
- Wash the cells twice with 1 ml ice-cold PBS.
- Remove PBS and add 800 µl ice-cold lysis buffer/well (supplemented with 1:100 protease inhibitor cocktail fleshly prior to use, see Recipes).
- Transfer the plate to an ice bucket and incubate for 15 min under gentle agitation on a titer plate shaker.
- After 15 min. incubation, gently homogenize the cell lysates (hereafter called ‘Cell lysate’ sample) on each well using a micropipette tip and transfer the lysates to micro-centrifuge tubes.
- Centrifuge the samples (Cell lysate or Media, see steps A12 and A16) for 20 min at 18,000 x g (or for 30 min at 14,000 x g) 4 °C.
- Transfer equal volume of cleared supernatant (~750 µl) from the tubes (cleared Cell lysate or Media samples) to new micro-centrifuge tubes, put them on ice and discard the pellets.
- Carryover of sedimented material into the immunoprecipitation steps may result in high nonspecific background.
- If necessary, the cleared Cell lysate and Media samples can be frozen at -80 °C until used for immunoprecipitation. However, it is preferable to harvest cells (from step A13) and freeze the cell pellet rather than the cell lysate (step A17) to avoid protein degradation.
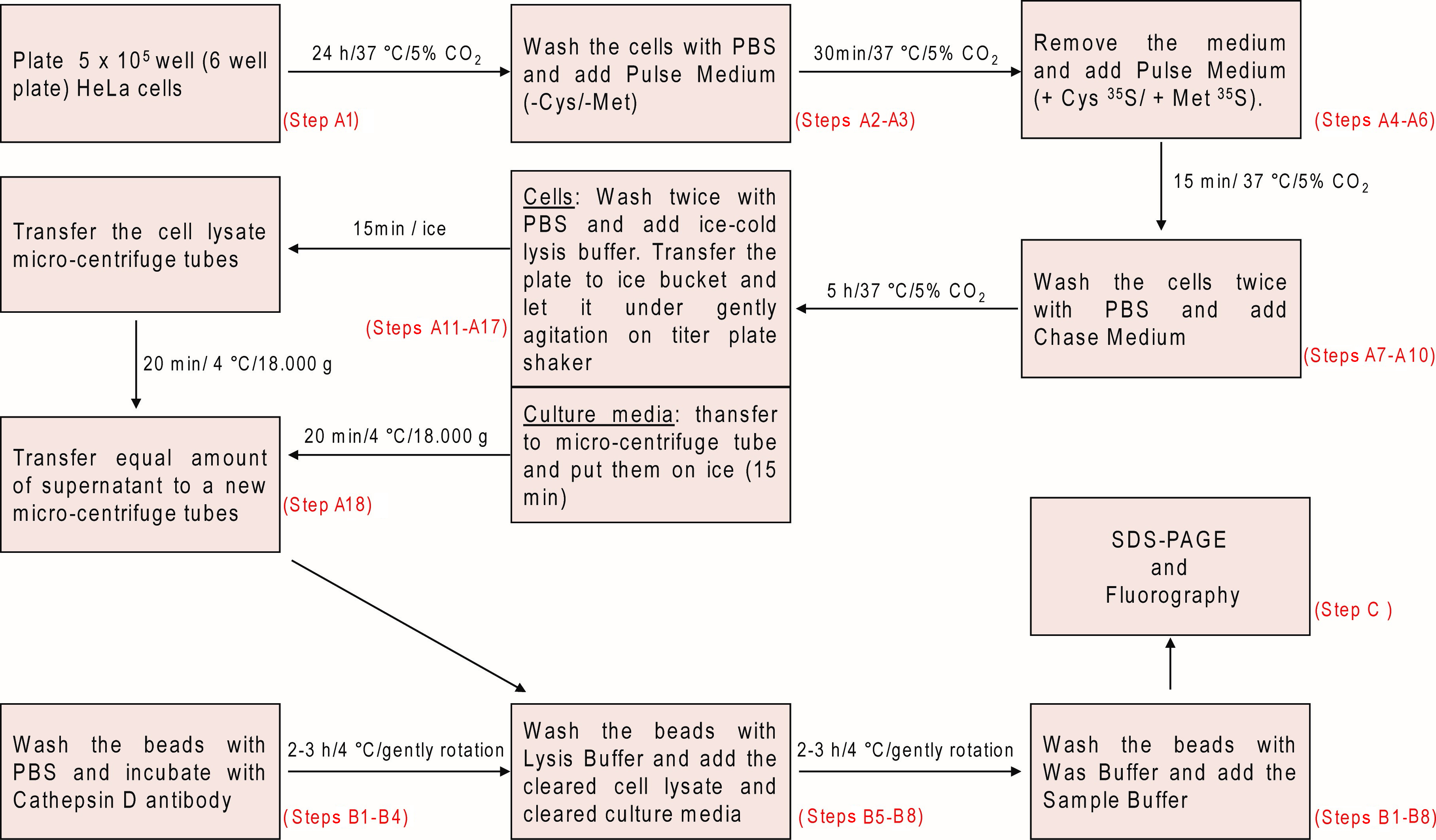
Figure 1. Schematic overview illustrating the metabolic labeling and pulse-chase analysis of Cathepsin D
- Carryover of sedimented material into the immunoprecipitation steps may result in high nonspecific background.
- Plate 5 x 105 HeLa CCL-2 cells/well (6-well plate) in 2 ml of complete DMEM and incubate for 24 h/37 °C/5% CO2 (Figure 1). Use one well for each time-point or condition.
- Immunoprecipitation
- Wash beads (30 μl of 50% Protein-A Ultralink resin/aqueous slurry per chase-time/condition point) 3 times with 500 μl of ice-cold PBS by centrifugation for 1 min at 3,000 x g, 4 °C (Figure 1). This step removes the aqueous slurry containing 0.02% sodium azide supplied with the beads, which might interfere with the antibody binding. Make sure to leave the initial volume of beads/slurry in the tube (~30 μl per chase-time/condition point).
- For large amounts of beads (more than 500 μl), a 15 ml conical polystyrene centrifuge tube may be used and washes are done with at least 1:3 volume of beads-slurry/PBS.
- It is important to start with around 10-20% extra volume of beads to compensate for loss during the wash and pipetting.
- For large amounts of beads (more than 500 μl), a 15 ml conical polystyrene centrifuge tube may be used and washes are done with at least 1:3 volume of beads-slurry/PBS.
- After washing, resuspend 30 μl of beads (in 50% PBS slurry) in 500 μl of ice-cold PBS containing 5 μl of 10% (v/v) BSA and 2 μl Cathepsin D antibody. Mix the suspension thoroughly.
When working with multiple samples, it is recommended to prepare a beads/antibody ‘master mix’. This is done by increasing the quantities indicated above proportionally to the number of samples, and incubating the suspension in a 15 ml conical polystyrene centrifuge tube. - Incubate for 2-3 h/4 °C under slow rotation on a tube rotator (capable of end-over-end inversion). During this step, the antibody will bind to the beads.
- Pellet the beads by centrifugation for 1 min at 3,000 x g, 4 °C and remove the supernatant.
- Wash the beads 3 times with 1 ml ice-cold lysis buffer by repeating step B4.
- After the final washing step, centrifuge the tubes containing the beads and remove the supernatant making sure to leave the initial volume of beads/slurry in the tube (~30 μl per chase-time/condition point).
- For each immunoprecipitation sample, incubate in a microcentrifuge tube: 30 μl beads/antibody (50% PBS slurry), 750 µl of cleared Cell lysate or Media, sample, 10 μl 10% BSA and 200 μl lysis buffer (supplemented with 1:100 protease inhibitor cocktail).
- Incubate for 2-3 h/4 °C under slow rotation on a tube rotator.
- After the incubation step, centrifuge the tubes, remove the supernatant and wash the beads 3 times each with 1 ml wash buffer (see Recipes) and one final wash step with 1 ml 1x PBS.
- Remove all PBS from the microcentrifuge tubes and resuspend the beads in 20 µl of 2x sample buffer (see Recipes). At this point, samples can be stored at -80 °C.
- Wash beads (30 μl of 50% Protein-A Ultralink resin/aqueous slurry per chase-time/condition point) 3 times with 500 μl of ice-cold PBS by centrifugation for 1 min at 3,000 x g, 4 °C (Figure 1). This step removes the aqueous slurry containing 0.02% sodium azide supplied with the beads, which might interfere with the antibody binding. Make sure to leave the initial volume of beads/slurry in the tube (~30 μl per chase-time/condition point).
- SDS-PAGE and fluorography
- Boil the samples for 5 min/95 °C.
- Spin the tubes and load the supernatant onto SDS-PAGE mini-gels (10%; 1 mm gels). Here you can also load a molecular weight standard on the gel.
- Run the electrophoresis for ~2 h at constant 120 V in running buffer (see Recipes). Note that running conditions and time will vary according to the electrophoresis system used.
- Fix gels with 25 ml/gel fixation solution (see Recipes) for 30 min at room temperature in a clean plastic container on titer plate shaker with slow shaking.
- Remove the fixation solution and incubate the gel with 25 ml/gel amplify fluorographic reagent for 30 min at room temperature on titer plate shaker with slow rotation.
- Dry gels onto chromatography paper in a gel dryer for 45 min/80 °C. Cover the gel with a plastic wrap to avoid sticking to the gel drier cover. The gel must be fully dehydrated (no longer sticky), 45 min at 80 °C is generally enough.
- Expose the BAS storage phosphor screen to the dried gel in an exposure cassette for generally 1 week at room temperature, protected from the light. The ideal exposure time will depend on the amount of labeled catD.
- Scan the screen using the Pharos FX plus molecular imager or equivalent and analyze the result by using the Quantity One 1-D Analysis Software (Bio-Rad Laboratories).
- Boil the samples for 5 min/95 °C.
Data analysis
After revealing the screen, the image should show bands indicative of the precursor Cathepsin D (pro-catD, ~52 kDa), the intermediate form (int-catD, ~48 kDa) and the mature lysosomal form (m-catD, ~34 kDa) in cell lysates and traces of pro-catD only in the media/supernatant sample (Figure 2). Note that these molecular masses vary slightly in different cell types. You can analyze the alterations in lysosomal sorting of cathepsin D by densitometric quantification of the different catD species. One way is to compare the intracellular abundance of precursor and mature forms with the abundance of secreted precursor, as the ratio of secreted to total catD, or to compare the abundance of mature form with the total catD as a ratio (Hirst et al., 2009). The relative abundance of each catD species can also be normalized by the number of methionine and cysteine residues present in the corresponding species (Kametaka et al., 2007). You can check the number of methionine and cysteine residues predicted for each human catD species in Minarowska et al., 2008.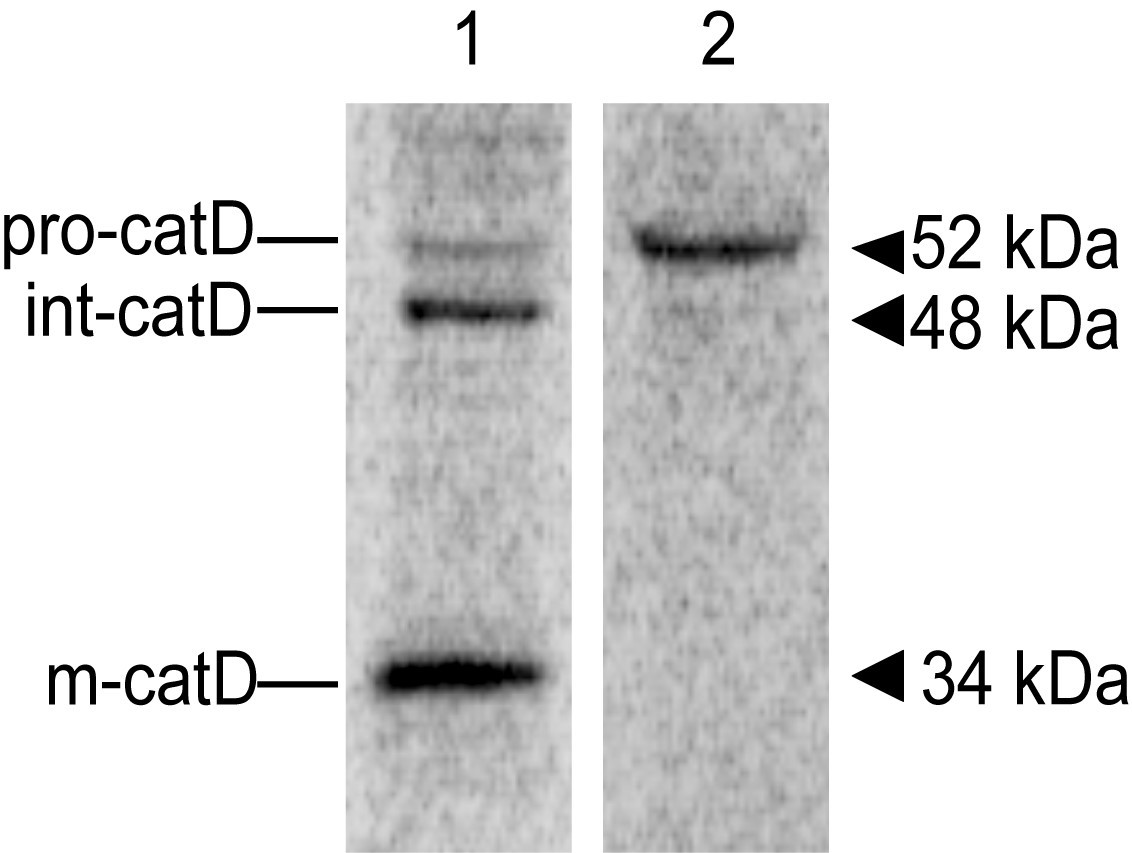
Figure 2. Representative image of a catD in vivo labeling/pulse-chase experiment. 24 h after plating into 6-well plates, HeLa cells were pulse-labeled with [35S] methionine-cysteine for 15 min at 37 °C, followed by incubation in chase buffer for 5 h at 37 °C. Cell lysate (lane 1) and Media (lane 2) samples were used for immunoprecipitation with an anti-cathepsin D antibody and the resulting immunoprecipitates were analyzed by SDS-PAGE and fluorography. The positions of pro-(pro-catD), intermediate (int-catD), and mature cathepsin D (m-catD) forms are indicated.
Recipes
- Dulbecco’s modified Eagle medium (DMEM) supplemented with 100 U of penicillin/ml and 0.1 mg/ml of streptomycin and10% (v/v) fetal bovine serum (FBS)
- 100x L-methionine/L-cysteine mix solution
- Dissolve 1.5 mg of both L-methionine and L-cysteine in 1 ml Milli-Q water
- Filter-sterilize in 0.22 µm syringe filter for storage
- Stored in a centrifuge tube at 4 °C
- Dissolve 1.5 mg of both L-methionine and L-cysteine in 1 ml Milli-Q water
- Pulse medium
Dulbecco’s modified Eagle’s medium methionine, cysteine and glutamine-free supplemented with:
2 mM L-glutamine
0.1 mCi/ml of [35S]-methionine-cysteine - Chase medium
Complete DMEM (Recipe 1) supplemented with 10 μl/ml of 100x L-methionine/L-cysteine mix solution - Lysis buffer (store at 4 °C)
50 mM Tris-HCl (pH 7.4)
300 mM NaCl
5 mM EDTA pH 8
0.5% (v/v) Triton X-100 - Wash buffer (store at 4 °C)
50 mM Tris-HCl (pH 7.4)
300 mM NaCl
5 mM EDTA pH 8
0.1% (v/v) Triton X-100 - 2x sample buffer
20 ml of 10% (v/v) SDS
16 ml of 0.5 M Tris-HCl pH 6.8
10 ml glycerol
0.1% bromophenol blue
4% β-mercaptoethanol
Milli-Q water
Note: This solution can be stored at room temperature but add β-mercaptoethanol fresh before use. - Fixation solution
50% methanol
10% acetic acid - Running buffer
30 g Tris
144 g glycine
10 g SDS
Complete with Milli-Q water for 1 L
Acknowledgments
This protocol has been adapted from a previously published paper (Tavares et al., 2017). This work was supported by the São Paulo Research Foundation (FAPESP) grant (2014/25812-0) to LLPdS and a PhD scholarship (2016/18207-9) to LAT. The authors declare no conflict of interest.
References
- Benes, P., Vetvicka, V. and Fusek, M. (2008). Cathepsin D--many functions of one aspartic protease. Crit Rev Oncol Hematol 68(1): 12-28.
- Bewley, M. A., Pham, T. K., Marriott, H. M., Noirel, J., Chu, H. P., Ow, S. Y., Ryazanov, A. G., Read, R. C., Whyte, M. K., Chain, B., Wright, P. C. and Dockrell, D. H. (2011). Proteomic evaluation and validation of cathepsin D regulated proteins in macrophages exposed to Streptococcus pneumoniae. Mol Cell Proteomics 10(6): M111 008193.
- Ghosh, P., Dahms, N. M. and Kornfeld, S. (2003). Mannose 6-phosphate receptors: new twists in the tale. Nat Rev Mol Cell Biol 4(3): 202-212.
- Hirst, J., Sahlender, D. A., Choma, M., Sinka, R., Harbour, M. E., Parkinson, M. and Robinson, M. S. (2009). Spatial and functional relationship of GGAs and AP-1 in Drosophila and HeLa cells. Traffic 10(11): 1696-1710.
- Kametaka, S., Moriyama, K., Burgos, P. V., Eisenberg, E., Greene, L. E., Mattera, R. and Bonifacino, J. S. (2007). Canonical interaction of cyclin G associated kinase with adaptor protein 1 regulates lysosomal enzyme sorting. Mol Biol Cell 18(8): 2991-3001.
- Minarowska, A., Gacko, M., Karwowska, A. and Minarowski, L. (2008). Human cathepsin D. Folia Histochem Cytobiol 46(1): 23-38.
- Poole, A. R., Dingle, J. T. and Barrett, A. J. (1972). The immunocytochemical demonstration of cathepsin D. J Histochem Cytochem 20(4): 261-265.
- Tavares, L. A., da Silva, E. M., da Silva-Januario, M. E., Januario, Y. C., de Cavalho, J. V., Czernisz, E. S., Mardones, G. A. and daSilva, L. L. (2017). CD4 downregulation by the HIV-1 protein Nef reveals distinct roles for the γ1 and γ2 subunits of the AP-1 complex in protein trafficking. J Cell Sci 130(2): 429-443.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Tavares, L. A. and daSilva, L. L. (2017). Monitoring the Targeting of Cathepsin D to the Lysosome by Metabolic Labeling and Pulse-chase Analysis. Bio-protocol 7(21): e2598. DOI: 10.21769/BioProtoc.2598.
Category
Biochemistry > Protein > Labeling
Biochemistry > Protein > Immunodetection
Molecular Biology > Protein > Detection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link