- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Determination of NO and CSF Levels Produced by Bacillus subtilis
Published: Vol 7, Iss 13, Jul 5, 2017 DOI: 10.21769/BioProtoc.2379 Views: 9240
Reviewed by: Jyotiska ChaudhuriSanjib Kumar GuhaTugsan Tezil

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

An Inexpensive Imaging Platform to Record and Quantitate Bacterial Swarming
Weijie Chen [...] Jay X. Tang
Sep 20, 2021 3635 Views

Purification of the Bacterial Amyloid “Curli” from Salmonella enterica Serovar Typhimurium and Detection of Curli from Infected Host Tissues
Murugesan Sivaranjani [...] Aaron P. White
May 20, 2022 3233 Views

A Guideline for Assessment and Characterization of Bacterial Biofilm Formation in the Presence of Inhibitory Compounds
Bassam A. Elgamoudi and Victoria Korolik
Nov 5, 2023 3102 Views
Abstract
The cell-to-cell communication and division of labour that occurs inside a beneficial biofilm produce significant differences in gene expression compared with the gene expression pattern of cells grew under planktonic conditions. In this sense, the levels of NO (nitric oxide) and CSF (Competence Sporulation Stimulating Factor) produced in Bacillus subtilis cultures have been measured only under planktonic growth conditions. We sought to determine whether NO and/or CSF production is affected in B. subtilis cells that develop as a biofilm. To measure the production levels of the two prolongevity molecules, we grew B. subtilis cells under planktonic and biofilm supporting condition.
Keywords: Bacillus subtilisBackground
NO is a key signalling molecule, playing a role in a variety of biological processes in vertebrates (Kerwin et al., 1995). C. elegans is unable to produce its own NO but is able to incorporate the NO produced by B. subtilis (Cabreiro and Gems, 2013; Gusarov et al., 2013; Kim, 2013; Clark and Hodgkin, 2014). Most organisms produce NO through aerobic conversion of L-arginine to L-citrulline in a reaction catalysed by the enzyme NO synthetase encoded by the nos gene (Sudhamsu and Crane, 2009). E. coli strains, several of which are routinely used to feed worms (OP50, HB101) (Cabreiro and Gems, 2013; Kim, 2013; Clark and Hodgkin, 2014), are not proficient in aerobic NO production because they lack a functional copy of nos (Sudhamsu and Crane, 2009). However, E. coli can produce NO under anaerobic/microaerophilic conditions by a series of biochemical reactions associated with the anaerobic respiratory chain of the bacterium (Corker and Poole, 2003). In such case, E. coli might find permissive conditions for NO production in the oxygen-depleted environment of the worm intestine. Bacteria produced-NO in worm gut that freely diffuses through the plasma membrane is oxidized to nitrate and nitrite, and thus, the concentration of nitrate and nitrite are directly proportional to the level of NO production (Gusarov et al., 2013) and can be determined using a colorimetric assay.
Intra- and interspecific quorum sensing (QS) constitutes molecules that bacteria use in nature to communicate with each other and with cells of different kingdoms (Shapiro, 1998; Ben Jacob et al., 2004; Parsek and Greenberg, 2005; Bassler and Losick, 2006). B. subtilis QS pentapeptide CSF (Competence Sporulation Stimulating Factor, also named PhrC) (Lazazzera et al., 1997) was previously reported to contribute to intestinal homeostasis by activating key survival pathways of the host (p38 MAP kinase and protein kinase B) and by inducing cytoprotective heat shock proteins (Hsps) (Fujiya et al., 2007; Willians, 2007). These effects of CSF (Willians, 2007) depend on its uptake by the protein OCTN2, a host cell membrane transporter of organic cations present in the apical face of epithelial cells (Fujiya et al., 2007). To quantify bacteria-produced CSF, promoters are commonly fused to heterologous reporter genes that encode enzymes that can be quantified using highly sensitive assays. Typically, incorporation to B. subtilis of a reporter lacZ gene, encoding β-galactosidase (β-gal) to the promoter region of a gene of interest, can be used to determine the level of CSF produced by this probiotic bacterium. When used in this manner, the Pcsf-lacZ fusion is integrated into the chromosome at the non-essential amyE locus. The basic colorimetric assay described here is the simplest and less expensive assay for quantifying β-gal activity. The cells are lysed and an aliquot of the extract is mixed with the reaction substrate, O-nitrophenyl-β-D-galactopyranoside (ONPG). When the yellow product becomes visible, the optical densities of the samples are determined spectrophotometrically.
Materials and Reagents
- Pipette tips
- Petri dishes 60 x 15 mm 500/cs (Fisher Scientific, catalog number: FB0875713A )
- Sterile 150 x 20 mm-culture tube (Sigma-Aldrich, catalog number: C1048 )
- Sterile 150 x 16 mm-culture tube (Science Lab Supplies, catalog number: 6135-5-012 )
- 1.5 ml tube
- Amicon Ultra 0.5 ml centrifugal filters MWCO 10 kDa (Sigma-Aldrich, catalog number: Z677108)
Manufacture: EMD Millipore, catalog number: UFC501096 . - Amicon Ultra centrifugal filter units Ultra-4, MWCO 30 kDa (Sigma-Aldrich, catalog number: Z648035)
Manufacture: EMD Millipore, catalog number: UFC803024 . - Cryovial (Simport, catalog number: T310-2A )
- 96-well solid plate (Colorimetric assay) (Cayman Chemical, catalog number: 400014 )
- 96-well cover sheet (Cayman Chemical, catalog number: 400012 )
- Derivative of B. subtilis NCIB31610
- Bacillus subtilis NCIB3610 and JH642 (Bacillus Genetic Stock Center, catalog numbers: 3A1 and 1A96 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S7653 )
- Bacto peptone (BD, BactoTM, catalog number: 211677 )
- Nitrate/Nitrite Colorimetric Assay Kit (Cayman Chemical, catalog number: 780001 )
- Nitrate/Nitrite assay buffer (Cayman Chemical, catalog number: 780022 )
- Nitrate Reductase Enzyme Preparation (Cayman Chemical, catalog number: 780010 )
- Nitrate Reductase Cofactor Preparation (Cayman Chemical, catalog number: 780012 )
- Nitrate Standard (Cayman Chemical, catalog number: 780016 )
- Griess Reagent R1 (Cayman Chemical, catalog number: 780018 )
- Griess Reagent R2 (Cayman Chemical, catalog number: 780020 )
- Luria Bertani broth (Sigma-Aldrich, catalog number: L3522 )
- Luria Bertani broth with agar (Sigma-Aldrich, catalog number: L2897 )
- Agar (Sigma-Aldrich, catalog number: A1296 )
- Potassium phosphate dibasic (K2HPO4) (Sigma-Aldrich, catalog number: P2222 )
- Potassium phosphate monobasic (KH2PO4) (Sigma-Aldrich, catalog number: P9791 )
- MOPS (Sigma-Aldrich, catalog number: M9381 )
- Magnesium sulfate heptahydrate (MgSO4·7H2O) (Sigma-Aldrich, catalog number: M1880 )
- Calcium chloride dihydrate (CaCl2·2H2O) (Sigma-Aldrich, catalog number: C3881 )
- Manganese(II) chloride (MnCl2·2H2O) (EMD Millipore, catalog number: 1059340100 )
- Glycerol (Sigma-Aldrich, catalog number: G5516 )
- Iron(III) chloride hexahydrate (FeCl3·6H2O) (Sigma-Aldrich, catalog number: 236489 )
- Glutamate (Sigma-Aldrich, catalog number: 49621 )
- Tryptophan (Sigma-Aldrich, catalog number: T0254 )
- Phenylalanine (Sigma-Aldrich, catalog number: P2126 )
- Chloramphenicol (Sigma-Aldrich, catalog number: C0378 )
- 100% ethanol (Sigma-Aldrich, catalog number: E7023 )
- Cholesterol (Sigma-Aldrich, catalog number: C8667 )
- Sodium phosphate dibasic (Na2HPO4) (Sigma-Aldrich, catalog number: S3264 )
- Sodium phosphate monobasic (NaH2PO4) (Sigma-Aldrich, catalog number: S3139 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9541 )
- β-mercaptoethanol (Sigma-Aldrich, catalog number: M6250 )
- Lysozyme from chicken egg white (Sigma-Aldrich, catalog number: L6876 )
- Triton X-100 (Sigma-Aldrich, catalog number: X100 )
- O-nitrophenyl-β-D-galactopyranoside (Sigma-Aldrich, catalog number: N1127 )
- Sodium carbonate (Na2CO3) (Sigma-Aldrich, catalog number: 223484 )
- 10 N NaOH
- Nematode growth medium (NGM) broth (see Recipes)
- MSgg medium (see Recipes)
- 5 mg/ml chloramphenicol (see Recipes)
- 5 mg/ml cholesterol (see Recipes)
- 1 M MgSO4 (see Recipes)
- 1 M CaCl2 (see Recipes)
- Phosphate buffer (see Recipes)
- Z buffer (see Recipes)
- 10 mg/ml lysozyme solution (see Recipes)
- 10% Triton X-100 solution (see Recipes)
- O-nitrophenyl-β-D-galactopyranoside (ONPG) 4.5 mg/ml (see Recipes)
- 1.2 M Na2CO3 (see Recipes)
- 50% glycerol (see Recipes)
- 100 mM MOPS pH = 7 (see Recipes)
Equipment
- Erlenmeyer flask (Fisher Scientific, catalog number: FB5006000 )
- Pipettor (Gilson, catalog number: F167300 )
- Glass pipette
- Refrigerated incubator (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 51028064 ; Fisher Scientific, catalog number: 37-20 )
Note: The product “Fisher Scientific, catalog number: 37-20 ” has been discontinued. - Water bath (AQUA® LYTIC incubator 37 °C)
- Freezers (-20 °C; So-Low Environmental Equipment) (Siemens, model: So-Low Ultra C85-22 )
- Autoclave (Tuttnauer, model: Model 6690 )
- Stirring hotplate (Corning, catalog number: 6795-620 )
- Centrifuge (Eppendorf, model: 5430 )
- Tabletop centrifuge (Eppendorf, model: 5424 )
- Molecular Dynamics Model SpectraMAX 340 PC microplate reader (Molecular Devices, model: SpectraMAX 340PC384 )
Procedure
- Preparation of B. subtilis strains culture
- Using sterile procedures, add 2 ml of sterile MSgg or NGM broth (see Recipes) in a sterile 150 x 20 mm-tube. Inoculate medium with 20 µl of a B. subtilis strains frozen culture (JH642 or NCIB3610) (see Note 3). For NGM broth’s preparation, mix 3 g NaCl and 2.5 g Bacto peptone in an Erlenmeyer flask. Add to 1 L of dH2O. Autoclave at 121 °C for 20 min. Cool flask in 55 °C water bath for 15 min. Add 1 ml 1 M CaCl2, 1 ml 5 mg/ml cholesterol in ethanol, 1 ml 1 M MgSO4 and 25 ml 1 M KPO4 buffer. Swirl to mix well.
- Incubate for 16-18 h at 37 °C with shaking (overnight culture).
- Using sterile procedures, add 2 ml of sterile MSgg or NGM broth (see Recipes) in a sterile 150 x 20 mm-tube. Inoculate medium with 20 µl of a B. subtilis strains frozen culture (JH642 or NCIB3610) (see Note 3). For NGM broth’s preparation, mix 3 g NaCl and 2.5 g Bacto peptone in an Erlenmeyer flask. Add to 1 L of dH2O. Autoclave at 121 °C for 20 min. Cool flask in 55 °C water bath for 15 min. Add 1 ml 1 M CaCl2, 1 ml 5 mg/ml cholesterol in ethanol, 1 ml 1 M MgSO4 and 25 ml 1 M KPO4 buffer. Swirl to mix well.
- Preparation of B. subtilis strains culture supporting biofilm formation
- Using sterile procedures, add 2 ml of sterile MSgg or NGM broth in a sterile 150 x 16 mm-tube. Inoculate medium with 20 µl of a B. subtilis strain overnight culture.
- Incubate at 25 °C for 36 h with (planktonic condition) or without shaking (biofilm-supporting condition) (see Note 4).
- Using sterile procedures, add 2 ml of sterile MSgg or NGM broth in a sterile 150 x 16 mm-tube. Inoculate medium with 20 µl of a B. subtilis strain overnight culture.
- Preparation of B. subtilis strains samples for NO determination (according to the kit’s instructions)
- Take the tubes and disrupt the biofilm by vortexing until a homogenous suspension is obtained. Take a 100 µl aliquot for CFU (colony-forming units) counting (see Note 1).
- Centrifuge at 12,470 x g for 20 min. Transfer the supernatant to a sterile 1.5 ml Eppendorf tube and discard pellet.
- Ultrafilter using a 10 or 30 kDa molecular weight cut-off filter using a commercially available centrifuge. The filters should be pre-rinsed with ultrapure water prior ultrafiltration.
- Wait for 3 h, the time required for the conversion of nitrate to nitrite.
- Take the tubes and disrupt the biofilm by vortexing until a homogenous suspension is obtained. Take a 100 µl aliquot for CFU (colony-forming units) counting (see Note 1).
- Preparation of a standard curve (according to the kit’s instructions)
- Standard curve for nitrate and nitrite must be included. If you plan to measure only total NO products (nitrate + nitrite), only the nitrate standard curve is required.
- In a clean tube, place 0.9 ml of ‘Nitrate Assay Buffer’ from the kit. To this, add 0.1 ml of reconstituted ‘Nitrate Standard’ and vortex for 20 sec. The concentration of this stock standard is 200 µM.
- Use this standard for the preparation of the nitrate standard curve as described below. The standard curve for nitrate is prepared by addition of reagents to the plate wells in the following way (Figure 1):
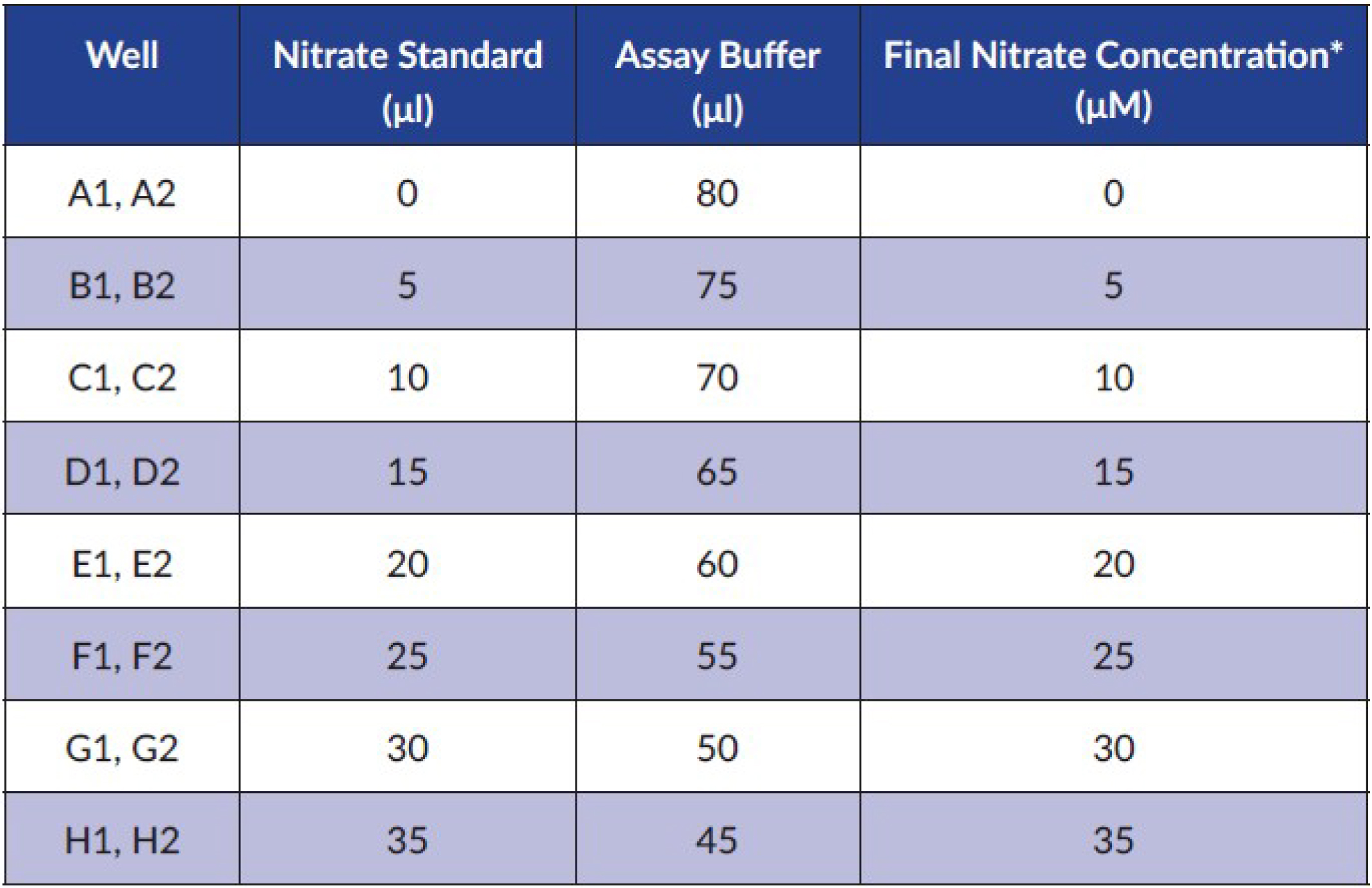
Figure 1. Preparation of nitrate standard curve according to the kit’s instructions
- Standard curve for nitrate and nitrite must be included. If you plan to measure only total NO products (nitrate + nitrite), only the nitrate standard curve is required.
- Performing the assay (according to the kit’s instructions)
- Add 200 µl of water or ‘Nitrate Assay Buffer’ to the blank wells. Do not add any other reagents to these wells.
- Add up to 80 µl of sample to the wells in a pattern you choose. The final volume must be adjusted to 80 µl using the ‘Nitrate Assay Buffer’ solution.
- Add 10 µl of the ‘Enzyme Cofactor Mixture’ to each of the wells (standards and unknown) (see Note 6).
- Add 10 µl of the ‘Nitrate Reductase Mixture’ to each of the wells (standards and unknown).
- Cover the plate with the plate cover and incubate at room temperature for 2 h. It is not necessary to shake the plate during incubation.
- After incubation time, add 50 µl of the ‘Griess Reagent R1’ to each of the wells (standards and unknown) (see Note 7). Pipette up and down 2-3 times.
- After incubation time, add 50 µl of the ‘Griess Reagent R2’ to each of the wells (standards and unknown) (see Note 7). Pipette up and down 2-3 times.
- Allow the color to develop for 10 min at room temperature. It is not necessary to cover the plate.
- Read the absorbance at 540 nm using a plate reader.
- Add 200 µl of water or ‘Nitrate Assay Buffer’ to the blank wells. Do not add any other reagents to these wells.
- Calculations
- Subtract the absorbance value of the blank wells from the absorbance values of all the other wells.
- Make a plot of the absorbance at 540 nm as a function of nitrate concentration. The nitrate standard curve is used for determination of total nitrate + nitrite concentration. An example is shown in Figure 2.
- Calculate NO concentration as follows: (Nitrate + Nitrite) (µM) = ([A540 - y - intercept)/slope] x (200 µl/volume of sample used (µl)) x dilution
- According to the CFU obtained in step C1, normalize NO (µM) to 1 x 108 CFU.
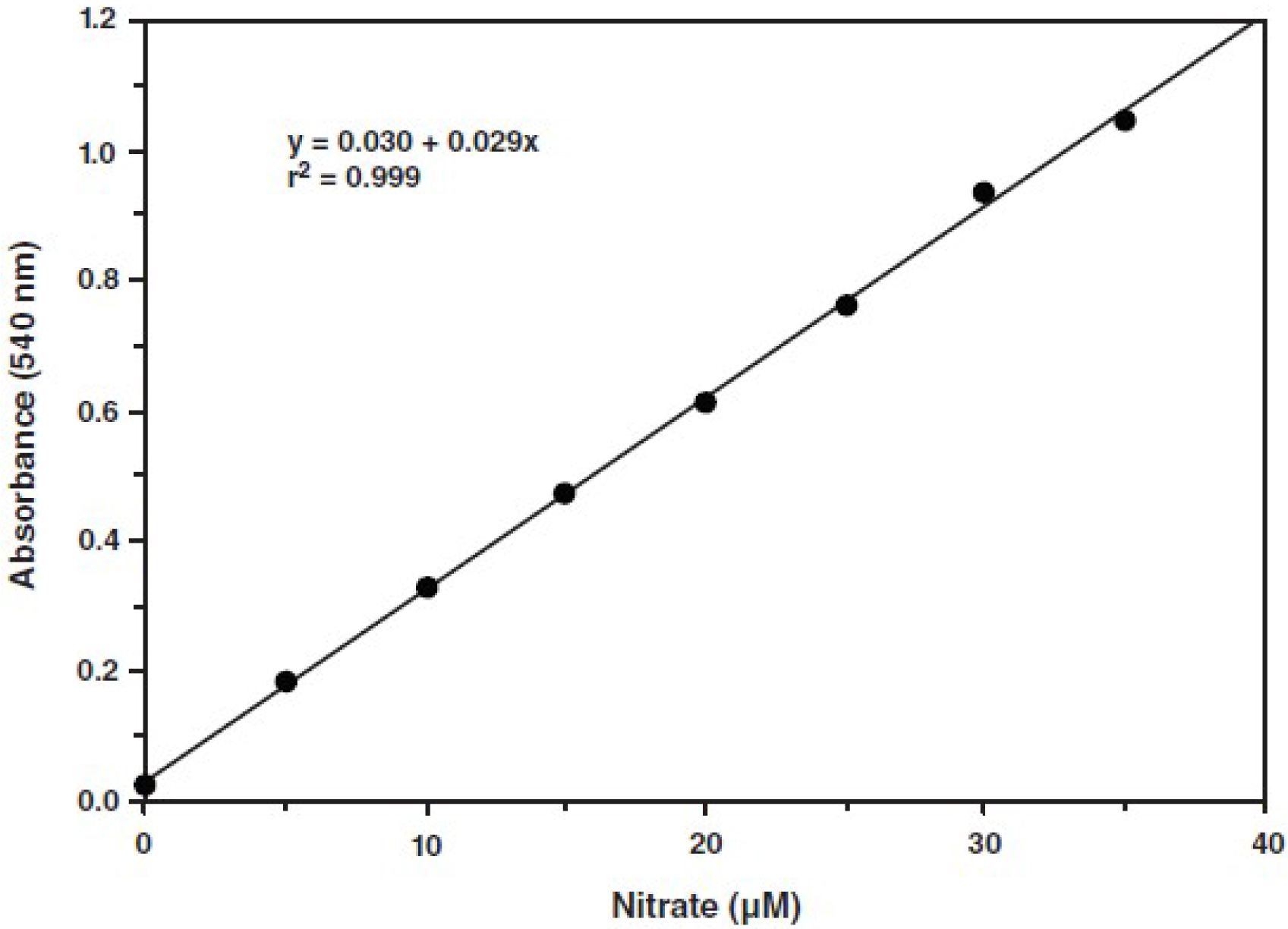
Figure 2. A typical standard curve (according to the kit’s instructions)
- Subtract the absorbance value of the blank wells from the absorbance values of all the other wells.
- Determination of β-galactosidase activity (Arabolaza et al., 2003) (see Note 5)
- The bacterial strain used in this study is a derivative of B. subtilis NCIB31610 containing Pcsf-lacZ fusion integrated into the bacterial chromosome at the non-essential amyE locus.
- Introduce 2 ml of MSgg or NGM broth aseptically into previously autoclaved 150 x 16 mm-culture tube and 2 µl of 5 mg/ml chloramphenicol.
- Each tube is inoculated with 20 µl of an overnight culture; uninoculated tubes containing only MSgg or NGM broth serve as controls for cross-contamination.
- The cultures are placed on a rotating shaker at 130 rpm (planktonic conditions) or without shaking (biofilm-supporting conditions) and incubated at 37 °C.
- The tubes are incubated for at least 36 h. To determine the culture density, take the tubes and disrupt the biofilm by vortexing until a homogenous suspension is obtained. Take a 100 µl aliquot for CFU counting (see Note 1). The A525 of the diluted culture (1/10 dilution) is determined using a Molecular Dynamics Model SpectraMAX 340PC microplate reader. As discussed below, dilution of the culture is required because cell density is not proportional to A525 when the values are above 0.8-1.
- Two aliquots of 1 ml from the disrupted culture are arrested by the addition of 2 µl of 5 mg/ml chloramphenicol (see Recipes) using a repeater pipettor and the tubes are placed on ice. Then, centrifuge the 1.5 ml Eppendorf tubes, for 5 min at 4 °C at 12,470 x g. Supernatants are removed and pellets are either frozen at -20 °C or assayed immediately (see Note 2) (Figure 3).

Figure 3. Flowchart image showing the basic steps of culturing bacteria in planktonic or biofilm-supporting conditions and samples collection - For cell permeabilization, 0.73 ml of Z buffer (see Recipes) is dispensed into the pellet.
- A reagent blank containing 0.73 ml of Z buffer is run with the unknowns.
- Add 10 µl of 10 mg/ml fresh lysozyme solution (see Recipes) to the tubes and incubate for 15 min in a 37 °C water bath.
- Transfer the tubes to bench and add 10 µl of 10% Triton X-100 (see Recipes). Permeabilization is accomplished by aspirating and dispensing the mixtures 10 times.
- Immediately after permeabilization, at zero time, the assay is initiated by adding 100 µl of 4.5 mg/ml ONPG (see Recipes) to each tube. In the end of the assay, the tubes are incubated at 28 °C for the appropriate length of time, e.g., 15 min, before the reaction is terminated by the addition of 150 µl of 1.2 M Na2CO3 to each tube (see Recipes).
- Then, the tubes’ content is transferred to a microplate, which is introduced into the plate reader and the A420 values are determined.
- Calculation of β-galactosidase activity expressed as Miller units (M.U.) is performed as follows:
M.U. = (OD420 x 66.7)/(OD525 x ml used)
where,
OD420 is the optical density of β-gal assay,
OD525 is the optical density of the bacterial culture,
66.7 is a correction factor,
ml used is the volume of Z buffer used to resuspend the bacterial pellet (in this case 0.73 ml). - According to the UFC obtained in step C1, normalize β-gal (M.U.) to 1 x 108 UFC.
- The bacterial strain used in this study is a derivative of B. subtilis NCIB31610 containing Pcsf-lacZ fusion integrated into the bacterial chromosome at the non-essential amyE locus.
Data analysis
- The absorbance data for A420 and A525 are transferred to a Microsoft Excel spreadsheet and β-galactosidase specific activities in Miller units are calculated.
- Each assay should be repeated at least three times for quadruplicate.
- Use the Student’s t-test with a significance cut-off level of P < 0.05 for comparisons between two groups.
- Use the one-factor (ANOVA) variance analysis and correct by the post hoc Bonferroni test for multiple comparisons.
Notes
- For the LB plates used to seed Bacillus subtilis, we strongly recommend drying plates at 45 °C for 20 min or 37 °C for 40 min upside down before to drop the dilutions onto each plate to avoid the sliding movement of this bacterium in solid surface with water drops from condensation.
- Samples should be used immediately or store for 1 month at -20 °C.
- Bacteria on an LB agar plate can be stored at 4 °C for a few weeks. However, for long-term storage, it is recommended to make glycerol stocks. The addition of glycerol stabilizes the frozen bacteria, preventing damage to the cell membranes and keeping the cells alive. A glycerol stock of bacteria can be stored stably at -80 °C for many years. For this, prepare an overnight bacteria culture, add 500 μl of the overnight culture to 500 μl of 50% glycerol (see Recipes) in a 2 ml cryovial and gently mix. Freeze the glycerol stock tube at -80 °C. The stock is now stable for years, as long as it is kept at -80 °C. Subsequent freeze and thaw cycles will reduce shelf life. To recover bacteria from your glycerol stock, open the tube and use a sterile loop or pipette tip to scrape some of the frozen bacteria off of the top. Do not let the glycerol stock thaw. Always keep your glycerol stock on ice while procedure is carried out. Streak the bacteria onto an LB agar plate. Grow Bacillus subtilis overnight at 37 °C.
- For incubation in planktonic condition, tubes inoculated with the bacterial overnight culture should keep in a 45° position with shaking at 130 rpm. For incubation in a biofilm-supporting condition, tubes inoculated with the bacterial overnight culture should keep in a vertical position without shaking.
- The parameters have been worked out with Bacillus subtilis and results obtained with other bacteria may not be accurate.
- The ‘Nitrate Reductase Cofactor Mixture’ is light-sensitive. Protect from light. Keep on ice during use. Store at -20 °C when not in use.
- The ‘Griess Reagent R1’ and ‘Griess Reagent R2’ are light-sensitive. Protect from light, especially sunlight. Store at 4 °C when not in use.
Recipes
- Nematode growth medium (NGM) broth
- Dissolve 3 g NaCl and 2.5 g Bacto peptone to 1 L of dH2O
- Autoclave
- Store at room temperature
- MSgg medium
- Dissolve 17.4 g K2HPO4, 13.6 g KH2PO4, 20.9 g MOPS, 18.3 g MgSO4 heptahydrate, 11 g CaCl2 dihydrate, 3.1 g MnCl2 dihydrate, 40.2 ml glycerol, 0.6 g FeCl3 hexahydrate, 20 g glutamate, 1 g tryptophan, 1 g phenylalanine to 100 ml of dH2O
- Autoclave
- Store at room temperature
- 5 mg/ml chloramphenicol
- Dissolve 0.05 g chloramphenicol to 10 ml of 100% ethanol
- Store at 4 °C
- 5 mg/ml cholesterol
- Dissolve 0.25 g of cholesterol in 50 ml of 100% ethanol
- Do not autoclave
- Store at room temperature
- 1 M MgSO4
- Dissolve 6 g MgSO4 heptahydrate in 50 ml of dH2O
- Autoclave
- Store at room temperature
- 1 M CaCl2
- Dissolve 5.55 g CaCl2 dihydrate in 50 ml of dH2O
- Autoclave
- Store at room temperature
- Phosphate buffer
- Dissolve 10.7 g K2HPO4 and 32.5 g KH2PO4 to 300 ml of dH2O
- Adjust pH to 6.0
- Autoclave
- Store at room temperature
- Z buffer
- Dissolve 2.55 g Na2HPO4, 1.44 g NaH2PO4, 0.22 g KCl, 0.074 MgSO4 heptahydrate and 1,050 µl β-mercaptoethanol to 300 ml of dH2O
- Store at room temperature
- 10 mg/ml lysozyme solution
- Dissolve 10 mg lysozyme to 1 ml of dH2O
- Store at 4 °C
- 10% Triton X-100 solution
- Dissolve 1 ml Triton X-100 to 10 ml of dH2O
- Store at room temperature
- O-nitrophenyl-β-D-galactopyranoside (ONPG) 4.5 mg/ml
- Dissolve 0.225 g ONPG to 50 ml of dH2O
- Store at -20 °C. Use until a yellow colour develops
- 1.2 M Na2CO3
- Dissolve 6.36 g Na2CO3 to 50 ml of dH2O
- Store at 4 °C
- 50% glycerol
- Dissolve 25 ml 100% glycerol to 25 ml of dH2O
- Autoclave
- Store at room temperature
- 100 mM MOPS pH = 7
- Dissolve 20.93 g MOPS in 80 ml of dH2O
- Adjust the pH to the desired value with 10 N NaOH
- Bring up the volume to 100 ml with dH2O
- Sterilize by filtration
Acknowledgments
This work was supported by CONICET (Consejo Nacional de Investigaciones Científicas y Técnicas) and FONCyT (Fondo para la Investigación Científica y Tecnológica) with the aid of the Pew Latin-American Program in Biological Sciences (Philadelphia, PA, USA), the Fulbright Committee (Washington, DC, USA) and former Fundación Antorchas (Buenos Aires, Argentina). We adapted the determination of β-galactosidase activity from Arabolaza et al. (2003).
References
- Arabolaza, A. L., Nakamura, A., Pedrido, M. E., Martelotto, L., Orsaria, L. and Grau, R. R. (2003). Characterization of a novel inhibitory feedback of the anti-anti-sigma SpoIIAA on Spo0A activation during development in Bacillus subtilis. Mol Microbiol 47(5): 1251-1263.
- Bassler, B. L. and Losick, R. (2006). Bacterially speaking. Cell 125(2): 237-246.
- Ben Jacob, E., Becker, I., Shapira, Y. and Levine, H. (2004). Bacterial linguistic communication and social intelligence. Trends Microbiol 12(8): 366-372.
- Cabreiro, F. and Gems, D. (2013). Worms need microbes too: microbiota, health and aging in Caenorhabditis elegans. EMBO Mol Med 5(9): 1300-1310.
- Clark, L. C. and Hodgkin, J. (2014). Commensals, probiotics and pathogens in the Caenorhabditis elegans model. Cell Microbiol 16(1): 27-38.
- Corker, H. and Poole, R. K. (2003). Nitric oxide formation by Escherichia coli. Dependence on nitrite reductase, the NO-sensing regulator Fnr, and flavohemoglobin Hmp. J Biol Chem 278(34): 31584-31592.
- Fujiya, M., Musch, M. W., Nakagawa, Y., Hu, S., Alverdy, J., Kohgo, Y., Schneewind, O., Jabri, B. and Chang, E. B. (2007). The Bacillus subtilis quorum-sensing molecule CSF contributes to intestinal homeostasis via OCTN2, a host cell membrane transporter. Cell Host Microbe 1(4): 299-308.
- Gusarov, I., Gautier, L., Smolentseva, O., Shamovsky, I., Eremina, S., Mironov, A. and Nudler, E. (2013). Bacterial nitric oxide extends the lifespan of C. elegans. Cell 152(4): 818-830.
- Kerwin, J. F., Jr., Lancaster, J. R., Jr. and Feldman, P. L. (1995). Nitric oxide: a new paradigm for second messengers. J Med Chem 38(22): 4343-4362.
- Kim, D. H. (2013). Bacteria and the aging and longevity of Caenorhabditis elegans. Annu Rev Genet 47: 233-246.
- Lazazzera, B. A., Solomon, J. M. and Grossman, A. D. (1997). An exported peptide functions intracellularly to contribute to cell density signaling in B. subtilis. Cell 89(6): 917-925.
- Parsek, M. R. and Greenberg, E. P. (2005). Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol 13(1): 27-33.
- Shapiro, J. A. (1998). Thinking about bacterial populations as multicellular organisms. Annu Rev Microbiol 52: 81-104.
- Sudhamsu, J. and Crane, B. R. (2009). Bacterial nitric oxide synthases: what are they good for? Trends Microbiol 17(5): 212-218.
- Willians, P. (2007). Bacillus subtilis: a shocking message from a probiotic. Cell Host Microbe 1(4): 248-249.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Cogliati, S., Rodriguez Ayala, F., Bauman, C., Bartolini, M., Leñini, C., Villalba, J. M., Argañaraz, F. and Grau, R. (2017). Determination of NO and CSF Levels Produced by Bacillus subtilis. Bio-protocol 7(13): e2379. DOI: 10.21769/BioProtoc.2379.
Category
Microbiology > Microbial signaling > Secondary messenger
Microbiology > Microbial biofilm > Biofilm culture
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link

















