- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Heparan Sulfate Identification and Characterisation: Method II. Enzymatic Depolymerisation and SAX-HPLC Analysis to Determine Disaccharide Composition
Published: Vol 7, Iss 7, Apr 5, 2017 DOI: 10.21769/BioProtoc.2197 Views: 9540
Reviewed by: Vivien Jane Coulson-Thomas Masahiro MoritaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Surface Plasmon Resonance for the Interaction of Capsular Polysaccharide (CPS) With KpACE
Zhe Wang [...] Chao Cai
Jun 20, 2025 3552 Views

New Approach to Detect and Isolate Rhamnogalacturonan-II in Arabidopsis thaliana Seed Mucilage
Dayan Sanhueza and Susana Saez-Aguayo
Sep 5, 2025 1240 Views

Detailed Method for the Purification of Rhamnogalacturonan-I (RG-I) in Arabidopsis thaliana
Liang Zhang [...] Breeanna R. Urbanowicz
Feb 5, 2026 99 Views
Abstract
Heparan sulfate (HS) is purified from complex matrices and often not fully characterised to validate its assignment. The characterisation of heparins and heparan sulfates through enzymatic depolymerisation and subsequent strong anion-exchange high performance liquid chromatography (SAX-HPLC) analysis and quantitation of the resulting disaccharides is a critical tool for assessing the structural composition of this class of compound. This protocol details a methodology to reproducibly determine the disaccharide composition of heparan sulfate by enzymatic depolymerisation and SAX-HPLC analysis. A complementary method for identification and characterisation of heparan sulfate can be found at Carnachan and Hinkley (2017).
Keywords: Heparan sulfateBackground
A number of methods exist for the structural analysis of heparin and HS. This protocol aims to provide an optimised methodology for the enzymatic depolymerisation of heparin and HS and the analysis and quantification of the disaccharides produced therein. Very few published analyses consider all aspects of the gross composition including the extent of depolymerisation in conjunction with the disaccharide composition obtained (Skidmore et al., 2006 and 2010; Carnachan et al., 2016). This is particularly worrisome when the sample is subsequently utilised in a biological assay that is invariably dose-dependent. The enzymatic procedure described herein is the culmination of a detailed study investigating the conditions necessary for optimal enzyme activity and HS depolymerisation (Carnachan et al., 2016). This procedure is intended to provide a stepwise protocol suitable for a laboratory inexperienced in glycosaminoglycan (GAG) analysis.
Materials and Reagents
- Microcentrifuge tubes (1.5 ml) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 3456 )
- Syringe filters (0.22 μm) (hydrophilic PTFE) (MicroScience Hydraflow, catalog number: MS SF13HY022 )
- Glass HPLC vials (2 ml) (Thermo Fisher Scientific, catalog number: THC11090500 )
- Septum lids (Thermo Fisher Scientific, catalog number: THC11090500 )
- Heparan sulfate (from porcine mucosa) (Celsus Laboratories, catalog number: HO-03103 )
- Heparin lyase I (heparinase I or heparitinase III, EC 4.2.2.7, [0.5 IU]) (IBEX Technologies, catalog number: 50-010 )
- Heparin lyase II (heparinase II or heparitinase II, no EC number assigned, [0.5 IU]) (IBEX Technologies, catalog number: 50-011 )
- Heparin lyase III (heparinase III or heparitinase I, EC 4.2.2.8, [0.5 IU]) (IBEX Technologies, catalog number: 50-012 )
- Heparin disaccharide standards produced by the action of bacterial heparinase on high grade porcine heparin (Iduron, catalog numbers: HD001 - HD008 and HD010 - HD013 )
- Water (distilled) (Sartorius arium® pro UV ultrafiltered; > 18.5 MΩ)
- Bovine serum albumin (BSA) (Thermo Fisher Scientific, GibcoTM, catalog number: 30060727 )
- Na2HPO4·H2O (EMD Millipore, catalog number: 106586 )
- NaOAc (ACS) (EMD Millipore, catalog number: 1062680250 )
- CaOAc·xH2O (Sigma-Aldrich, catalog number: 25011 )
- Sodium chloride (NaCl) (99.99%) (EMD Millipore, Suprapur®, catalog number: 1.06406 )
- Hydrochloric acid (HCl) (37%) (EMD Millipore, catalog number: 100317 ) and NaOH (Sigma-Aldrich, catalog number: S8045 ) made up to 1 N aqueous solutions for adjusting pH
- Phosphoric acid (85%) (Sigma-Aldrich, catalog number: 79606 )
- Enzyme storage buffer (see Recipes)
- Digestion media (see Recipes)
- Mobile phases for HPLC analysis (see Recipes)
Equipment
- High performance liquid chromatography (HPLC) machine (Infinity with multiple wavelength detector) (Agilent Technologies, model: 1260 )
- Strong anion-exchange HPLC column (4 x 250 mm) (ProPacTM PA1, Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 039658 ) with guard column (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 039657 )
- Pipettors (0.5-10 μl, 20-200 μl and 100-1,000 μl) (Eppendorf)
- Incubator (BINDER, model: KBF-115 )
- Rotator (Cole-Parmer, Stuart, model: SB3 )
- Centrifuge (Eppendorf, model: Minispin® plus , catalog number: 5453000011)
Procedure
- Digest procedure
- Enzyme solutions (50 µl) as received (-80 °C) are permitted to warm to RT then diluted to 500 µl with the appropriate storage buffer giving, nominally, 0.5 IU per 500 µl. Aliquots (5 µl) are stored frozen (-80 °C) until needed. These 5 µl aliquots contain 5 mIU of activity according to the supplier's specification. However, when the enzyme activities were assayed using Celsus heparan sulfate and the digestion conditions described below, they were found to be 2.8, 5.0 and 13.8 mIU for Heparin lyases (HL) I, II and III, respectively. HL activities should be independently verified prior to starting the depolymerisation procedure.
- Heparin or HS (1 mg, in duplicate) is dissolved in a microcentrifuge tube (1.5 ml) with the digestion media (470 µl).
- Sequential enzyme addition is completed.
- HL I (one 5 µl aliquot) is added and the solution incubated (37 °C, 2 h, inversion at 9 rpm).
- HL III (5 µl) is added and incubation continued (1 h).
- HL II (5 µl) is added and incubation continued (18 h).
- All three enzymes (an additional 5 µl of each) are added at the same time and the solution incubated (gentle inversion, 9 rpm) for a further 24 h.
- Digests are terminated by heating (100 °C, 5 min).
- Samples are centrifuged (14,000 x g, 10 min) and the supernatant recovered.
- The supernatants (2 mg/ml) are then accurately diluted with water to give 100 µg/ml solutions, by taking 50 µl of supernatant and adding 950 µl of water. The 100 µg/ml solutions are then filtered (0.22 µm) and analysed.
- Enzyme solutions (50 µl) as received (-80 °C) are permitted to warm to RT then diluted to 500 µl with the appropriate storage buffer giving, nominally, 0.5 IU per 500 µl. Aliquots (5 µl) are stored frozen (-80 °C) until needed. These 5 µl aliquots contain 5 mIU of activity according to the supplier's specification. However, when the enzyme activities were assayed using Celsus heparan sulfate and the digestion conditions described below, they were found to be 2.8, 5.0 and 13.8 mIU for Heparin lyases (HL) I, II and III, respectively. HL activities should be independently verified prior to starting the depolymerisation procedure.
- Analysis procedure
- HPLC is completed using a binary solvent system and a flow rate of 1 ml/min at 40 °C. Following long-term storage and prior to injecting the first sample, the column is conditioned by flushing thoroughly with water, then with 100% B for 10 min before equilibrating the column with the starting conditions (0% B) for 2 min. Digest and standard solutions (see below) are injected by autosampler (50 µl injection volume) and the disaccharides separated using a gradient system, 0% B (1 min), 0% to 35% B (over 31 min), 35% to 65% B (over 15 min), 100% B (10 min). Post analysis column re-equilibration 0% B (3 min).
- Disaccharides are detected by absorbance at their absorbance maxima of 232 nm.
- Standard curves for each disaccharide standard are prepared by dissolution of each of the twelve heparin-derived disaccharide standards (supplied as 1.0 mg samples) to give 1 mg/ml stock solutions in water. These stock solutions are in turn used to prepare a twelve disaccharide standard mix containing 20 µg/ml of each standard which was subsequently serially diluted to prepare solutions containing 10, 5, 2.5, 1.25, 0.625 and 0.3125 µg/ml of each disaccharide. These standard solutions are analysed with each batch of digests (see Figure 1). Linear calibration curves (concentration vs peak area) with R2 values of > 0.998 can be generated.
- For the enzyme digests, data reported are the average of two digests. Each digest is analysed by HPLC in duplicate (Figure 1). Disaccharides in the digest can be identified by elution times relative to the twelve heparin-derived disaccharide standards and quantified using the calibration curves.
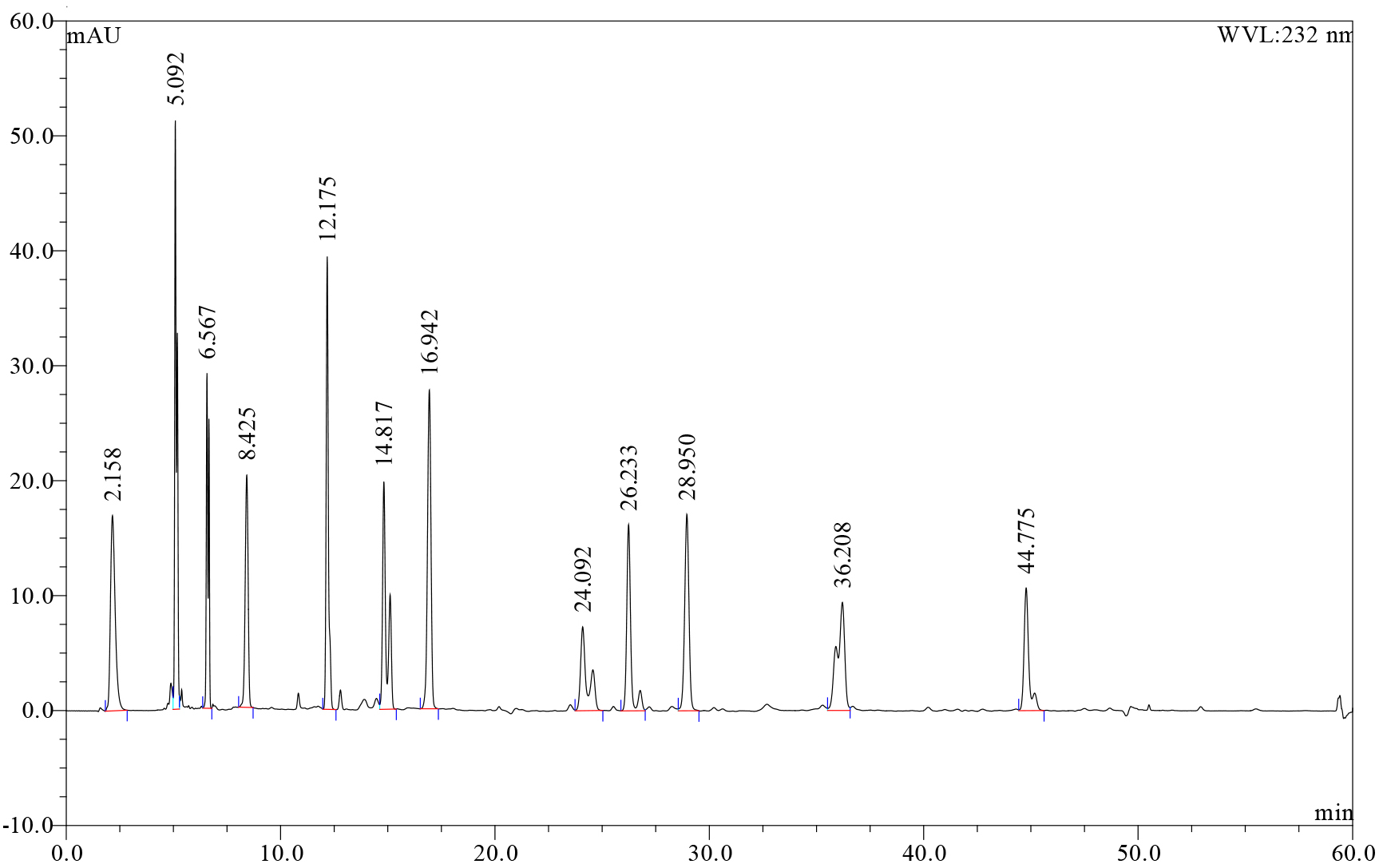
Figure 1. HPLC chromatogram showing resolution of the twelve heparin-derived disaccharides present in the standard mix. Retention times and species: 2.158 min, Δ-UA-GlcN; 5.092 min, Δ-UA-GlcNAc; 6.567 min, Δ-UA-GlcN(6S); 8.425 min, Δ-UA(2S)-GlcN; 12.175 min, Δ-UA-GlcNS; 14.817 min, Δ-UA-GlcNAc(6S); 16.942 min, Δ-UA(2S)-GlcNAc; 24.092 min, Δ-UA(2S)-GlcN(6S); 26.233 min, Δ-UA-GlcNS(6S); 28.950 min, Δ-UA(2S)-GlcNS; 36.208 min, Δ-UA(2S)-GlcNAc(6S); 44.775 min, Δ-UA(2S)-GlcNS(6S). The elution order of the twelve disaccharide standards was initially determined by running each species separately under the chromatographic conditions described above.
- HPLC is completed using a binary solvent system and a flow rate of 1 ml/min at 40 °C. Following long-term storage and prior to injecting the first sample, the column is conditioned by flushing thoroughly with water, then with 100% B for 10 min before equilibrating the column with the starting conditions (0% B) for 2 min. Digest and standard solutions (see below) are injected by autosampler (50 µl injection volume) and the disaccharides separated using a gradient system, 0% B (1 min), 0% to 35% B (over 31 min), 35% to 65% B (over 15 min), 100% B (10 min). Post analysis column re-equilibration 0% B (3 min).
Data analysis
- A typical HPLC chromatogram for an enzyme digest of heparan sulfate is displayed in Figure 2.
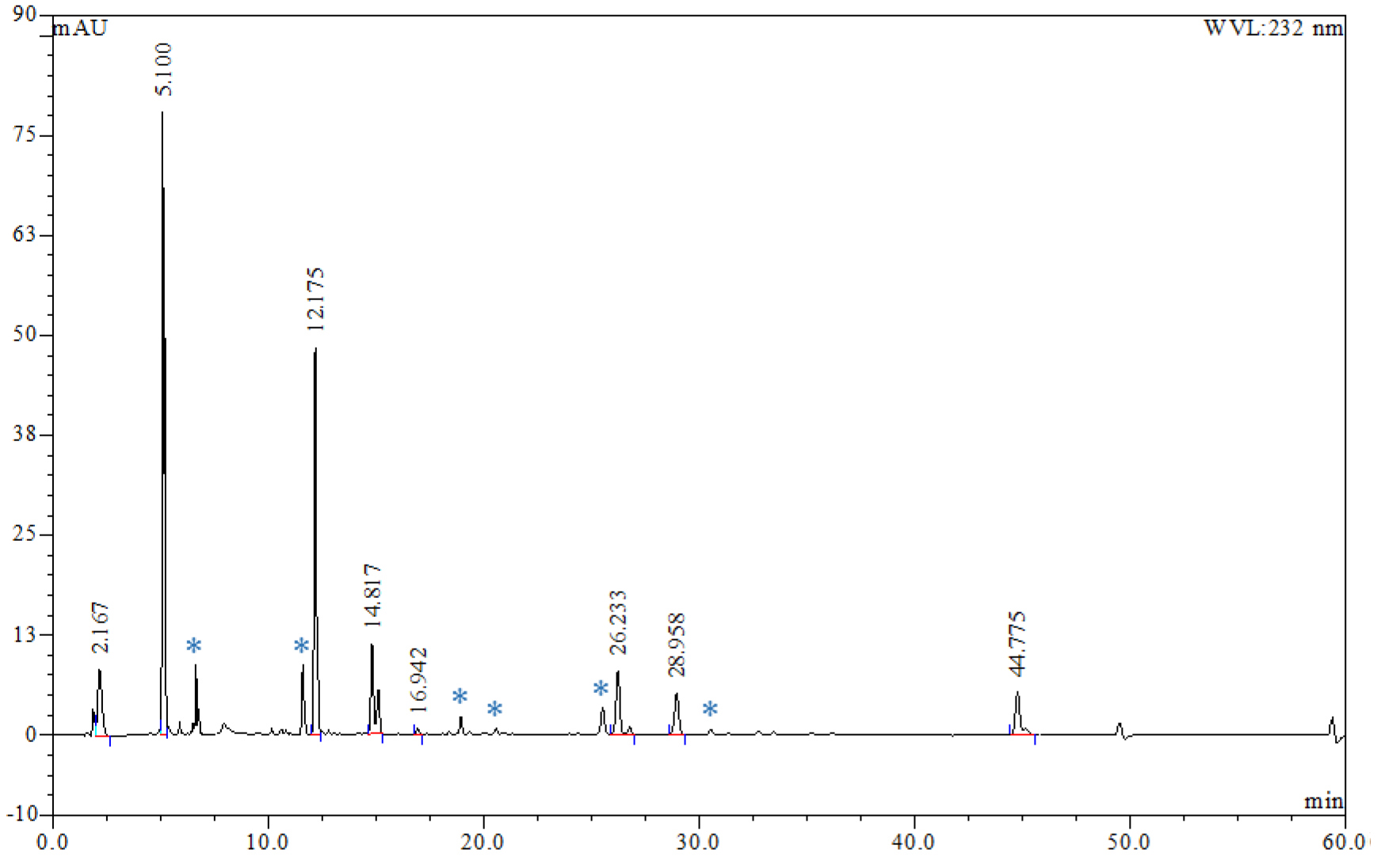
Figure 2. HPLC chromatogram of a typical enzymatic digest of commercial porcine mucosal heparan sulfate showing separation of the Δ-disaccharide constituents. Peaks marked with an asterisk represent uncharacterized species that appear reproducibly in chromatographic runs. - Using the standard curves and integrated areas for each of the disaccharide peaks it is possible to determine the proportion of each disaccharide present in the sample. This data can be used to calculate the normalised proportion of each disaccharide and the total mass of disaccharides produced by the digestion (Table 1).
- The compositions reported are the mean from the four HPLC runs; duplicate analysis of the two digests completed on each sample.
Table 1. Disaccharide normalised composition and mass recovery for a commercial porcine mucosal heparan sulfate
Notes
- Enzyme activity is tested using the same conditions as above and using the reference HS specified in the materials and methods above.
- The split peaks observed for some disaccharides in the HPLC traces have been attributed to the two anomeric forms of the reducing sugar moiety. It is important to be consistent and either quantitate individual anomeric species (Beccati et al., 2016), or, as in this protocol, the peak areas are pooled for quantifying disaccharides from sample digests.
- The disaccharide with an N-unsubstituted amine group and no sulfation (ΔUA-GlcN) can be problematic to quantify on this ion-exchange column. Care should be taken to ensure that this species, which is poorly-retained on the column, is not co-eluting with the solvent front and providing erroneous values.
- Some additional peaks were reproducibly detected in the chromatograms of digests (see species labelled * on Figure 2). These are tentatively attributed to tetrasaccharides resulting from incomplete enzymatic depolymerisation but have not been characterised fully.
- The data generated using this SAX-HPLC method can be used to quantify the total mass of unsaturated disaccharides produced by heparin lyase depolymerisation and, in theory, should provide a guide as to the purity of the heparan sulfate. However, if heparin or specific linkages not susceptible to enzymatic depolymerisation are present, then the extent of depolymerisation determined using this method can be misleading in a purity calculation (Yamada et al., 1993; Carnachan et al., 2016; Mulloy et al., 2016).
- During the analysis of an unknown HS material, the reference HS described in this protocol (Celsus) is always digested and analysed in duplicate to ensure that the protocol is operating within expectations.
Recipes
- Enzyme storage buffer
Heparin lyases (HL) are stored with BSA (0.1%, w/v) in 50 mM sodium phosphate buffer prepared by the addition of Na2HPO4·H2O to RO water with adjustment of the pH by the addition of concentrated phosphoric acid to give pH 7.1 for HL I and II and pH 7.6 for HL III. 100 mM NaCl is added to the storage buffer for HL I only - Digestion media
NaOAc (100 mM) with CaOAc (2 mM) and adjustment of the pH to 7.0 by the addition of 1 N NaOH or HCl - Mobile phases for HPLC analysis
Water (eluent A) and 2 M NaCl (aqueous, eluent B) are both prepared and adjusted to pH 3.5 using 1 N HCl immediately prior to analysis
Acknowledgments
This research was supported in part by the New Zealand Ministry of Business, Innovation and Employment, and the Kiwi Innovation Network (KiwiNet, VL001298). The collaborative research completed with Drs Simon M. Cool, Victor Nurcombe and R. Alex A. Smith (Institute of Medical Biology, Agency for Science, Technology and Research, Immunos, Singapore) is acknowledged.
References
- Beccati, D., Lech, M., Ozug, J., Gunay, N. S., Wang, J., Sun, E. Y., Pradines, J. R., Farutin, V., Shriver, Z., Kaundinya, G. V. and Capila, I. (2016). An integrated approach using orthogonal analytical techniques to characterize heparan sulfate structure. Glycoconj J 34(1): 107-117.
- Carnachan, S. M., Bell, T. J., Sims, I. M., Smith, R. A., Nurcombe, V., Cool, S. M. and Hinkley, S. F. (2016). Determining the extent of heparan sulfate depolymerisation following heparin lyase treatment. Carbohydr Polym 152: 592-597.
- Carnachan, S. M. and Hinkley, S. F. R. (2017). Heparan Sulfate Identification and Characterisation: Method I. Heparan Sulfate Identification by NMR analysis. Bio-protocl 7(07): e2196.
- Mulloy, B., Wu, N., Gyapon-Quast, F., Lin, L., Zhang, F., Pickering, M. C., Linhardt, R. J., Feizi, T. and Chai, W. (2016). Abnormally high content of free glucosamine residues identified in a preparation of commercially available porcine intestinal heparan sulfate. Anal Chem 88(13): 6648-6652.
- Skidmore, M. A., Guimond, S. E., Dumax-Vorzet, A. F., Atrih, A., Yates, E. A. and Turnbull, J. E. (2006). High sensitivity separation and detection of heparan sulfate disaccharides. J Chromatogr A 1135 (1): 52-56.
- Skidmore, M. A., Guimond, S. E., Dumax-Vorzet, A. F., Yates, E. A. and Turnbull, J. E. (2010). Disaccharide compositional analysis of heparan sulfate and heparin polysaccharides using UV or high-sensitivity fluorescence (BODIPY) detection. Nat Protoc 5 (12): 1983-1992.
- Yamada, S., Yoshida, K., Sugiura, M., Sugahara, K., Khoo, K. H., Morris, H. R. and Dell, A. (1993). Structural studies on the bacterial lyase-resistant tetrasaccharides derived from the antithrombin III-binding site of porcine intestinal heparin. J Biol Chem 268(7): 4780-4787.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Carnachan, S. M. and Hinkley, S. F. R. (2017). Heparan Sulfate Identification and Characterisation: Method II. Enzymatic Depolymerisation and SAX-HPLC Analysis to Determine Disaccharide Composition. Bio-protocol 7(7): e2197. DOI: 10.21769/BioProtoc.2197.
Category
Biochemistry > Carbohydrate > Disaccharide
Biochemistry > Carbohydrate > Polysaccharide
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









