- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Evaluation of Burkholderia cepacia Complex Bacteria Pathogenicity Using Caenorhabditis elegans
Published: Vol 6, Iss 20, Oct 20, 2016 DOI: 10.21769/BioProtoc.1964 Views: 7695
Reviewed by: Peichuan ZhangAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Artificial Optogenetic TRN Stimulation of C. elegans
Ithai Rabinowitch [...] Jihong Bai
Oct 20, 2016 8620 Views

Testing for Assortative Mating by Diet in Drosophila melanogaster
Philip T Leftwich and Tracey Chapman
Oct 20, 2018 5667 Views

Aerotaxis Assay in Caenorhabditis elegans to Study Behavioral Plasticity
Qiaochu Li [...] Karl Emanuel Busch
Aug 20, 2022 2251 Views
Abstract
This protocol describes two biological assays to evaluate pathogenicity of Burkholderia cepacia complex (Bcc) strains against the nematode Caenorhabditis elegans. Specifically, these two assays allow one to identify if the under-investigated Bcc strains are able to kill the nematodes by intestinal colonization (slow killing assay, SKA) or by toxins production (fast killing assay, FKA). The principal differences between the two assays rely on the different killing kinetics for worms.
Keywords: Burkholderia cepacia complex strainsBackground
The Burkholderia cepacia complex (Bcc) occupies a critical position among Gram-negative multi-drug resistant bacteria. It consists of at least 20 closely related species. Many Bcc strains are multi drug and pandrug-resistant opportunistic human pathogens caused problematic lung infections in immune-compromised individuals, including cystic fibrosis (CF) patients. The use of non-vertebrate host model can be useful for dissecting virulence and pathogenicity determinants as well as identifying novel therapeutic targets (Kothe et al., 2003).
There are a good number of assays for detecting Bcc virulence against a large panel of host models, in liquid or in solid surface. However, some of those are mostly focused on phenotypic observations, which are difficult to detect and have a low reproducibility (Cardona et al., 2005). Herein, we developed two assays based on the analysis of surviving worms, which is a more reproducible and allows easy and fast comparison among the Bcc strains tested. In addition, these assays permit the detection of death mechanisms of Bcc towards nematode.
These killing assays allow us to identify bacterial strains that are able to colonize the nematode intestine and produce diffusible toxins capable of killing the host.
Materials and Reagents
- 15 ml Falcon tubes (Corning, Falcon®, catalog number: 352095 )
- 3.5 cm diameter agar plates (Corning, catalog number: 430165 )
- Caenorhabditis elegans
- Burkholderia cepacia complex (Bcc) strains
- E. coli OP50
- NaOH
- Bleach (Aurora)
- NGM agar
- PGS agar
- Sodium chloride (NaCl)
(Sigma-Aldrich, catalog number: S9888 )
- Tryptone
(Conda, catalog number: 1612 )
- Yeast extract (Conda, catalog number: 1702 )
- Peptone (Conda, catalog number: 1602 )
- European agar (Conda, catalog number: 1800 )
- Magnesium sulfate (MgSO4) (Sigma-Aldrich, catalog number: M7506 )
- Calcium chloride (CaCl2) (Sigma-Aldrich, catalog number: C1016 )
- Cholesterol (Sigma-Aldrich, catalog number: C3045 )
- Glucose (Conda, catalog number: 1900 )
- Sorbitol (Sigma-Aldrich, catalog number: S1876 )
- Potassium phosphate monobasic (KH2PO4)
(Sigma-Aldrich, catalog number: P5655 )
- Sodium phosphate dibasic (Na2HPO4)
(Sigma-Aldrich, catalog number: S5136 )
- LB broth (see Recipes)
- NGM agar medium (see Recipes)
- PGS agar medium (see Recipes)
- M9 buffer (see Recipes)
Equipment
- Centrifuge
- 20 °C chamber
- 37 °C shaking incubator
- Dissecting microscope
- Platinum loop
Software
- Graph-pad Prism 5 software
Procedure
- Day 1
- Synchronize worms with bleaching protocols: use C. elegans plates with many gravid hermaphrodites (Stiernagle, 2006). Wash the plates with sterile H2O.
- Collect the liquid in a sterile 15 ml Falcon tube. Add H2O to a total volume of 3.5 ml.
- Mix 0.5 ml 5 N NaOH with 1 ml bleach. Make this solution fresh just before use! Add to the centrifuge tube with the worms.
- Vortex the tube for a few seconds. Repeat vortexing every 2 min for a total of 10 min.
- Spin the tube in a centrifuge for 30 sec at 1,300 x g to pellet the released eggs.
- Aspirate to 0.1 ml.
- Add sterile H2O to 5 ml. Vortex for a few seconds.
- Repeat steps A6 and A7.
- Transfer the eggs in the remaining 0.1 ml of liquid to the edge of a clean NGM plate seeded with an E. coli OP50 lawn and incubate at 20 °C.
Note: If you use C. elegans mutants that do not change their intrinsic nature under high temperature, you can incubate eggs at 25 °C as some mutants grow slower than WT. - Inoculate Bcc strains in 15 ml Falcon tubes containing 3 ml of LB and incubate the tubes at 37 °C for 24 h in shaking condition (220 rpm).
Note: As controls, use E. coli OP50 in the place of Bcc strains.
- Synchronize worms with bleaching protocols: use C. elegans plates with many gravid hermaphrodites (Stiernagle, 2006). Wash the plates with sterile H2O.
- Day 2
- Normalize Bcc cultures at 1.7 OD/ml and seed 50 μl of the culture on 3.5 cm diameter plates containing 3 ml of NGM agar (slow killing assay, SKA) or PGS agar (fast killing assay, FKA). Incubate the plates O/N at 37 °C.
Note: Incubation of Bcc strains at 37 °C should never exceed 16 h. Do not store Bcc seeded plates at 4 °C, as many of these pathogens are very sensitive to temperature and this can cause variation. - Check the developmental stage of worms. After 24 h worms should be at larval stage L2 or L3 when grown at 20 °C.
- Normalize Bcc cultures at 1.7 OD/ml and seed 50 μl of the culture on 3.5 cm diameter plates containing 3 ml of NGM agar (slow killing assay, SKA) or PGS agar (fast killing assay, FKA). Incubate the plates O/N at 37 °C.
- Day 3
- Wash synchronized worms at larval stage 4 (L4) off plates with M9 buffer and collect them in 15 ml Corning tubes.
- Wash worms 2-3 times with M9 buffer to remove residual bacterial cells.
- Spot 30-40 L4 worms on the plates seeded with Bcc strains (5 replicas).
Note: Before adding the worms, plates should be kept at room temperature to cool them down after the incubation at 37 °C. - Count worms at time 0. Incubate the plates at 20 °C and perform daily count of surviving worms up to day 5 (for FKA) and day 6 (for SKA).
Note: A worm is considered dead when it no longer responds to gentle touch with a platinum wire. - At the end of the experiment, calculate the average percentage of surviving worms.
- Evaluate Bcc pathogenicity. The virulence ranking (VR) ranges from 0 to 3 and it is based on the percentage of surviving worms after the period of observation. A Bcc strain is considered to be non-pathogenic (VR = 0) when the percentage of worms alive at the conclusion of the period of observation ranges from 100% to 80%; VR = 1 corresponds to a percentage of worms alive between 79% to 50%; VR = 2 corresponds to a percentage of worms alive between 49% to 6%; finally, the VR is considered 3 when the percentage of surviving worms was ≤ 5% (Figure 1).
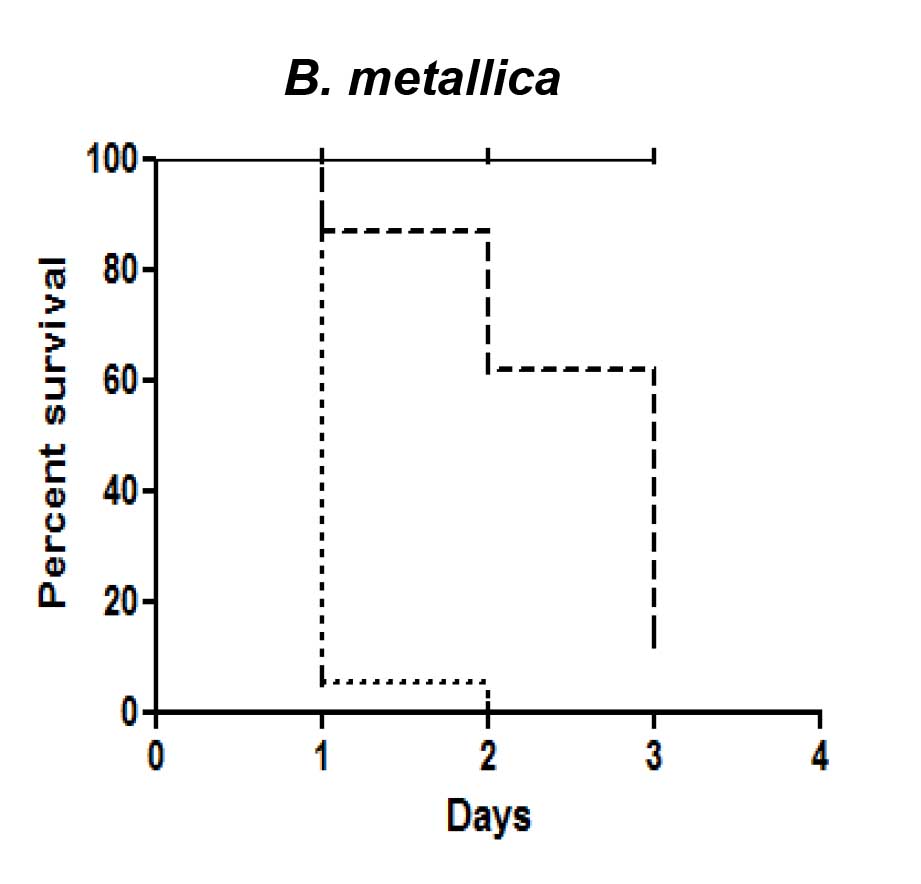
Figure 1. Kaplan-Meier survival plots for L4 stage WT worms fed with E. coli OP50 (solid lines), Burkholderia metallica on NGM (dashed lines), Burkholderia metallica on PGS (dotted lines). n: Number of worms at day 0. The pathogenicity of Bcc strain B. metallica on SKA (n = 80) was compared with the ability on FKA (n = 113).
- Wash synchronized worms at larval stage 4 (L4) off plates with M9 buffer and collect them in 15 ml Corning tubes.
Data analysis
Kaplan-Meier survival curves can be generated and analyzed using Graph-pad Prism 5 software. Comparisons vs. control for both the C. elegans are performed using Fisher’s exact test to account for possible non-Normality in the data. Bonferroni-Holm correction of P-values is used to account for the multiple comparisons performed. More details can be found in Tedesco et al. (2015).
Notes
- These protocols can be very useful for the identification of virulent Bcc strains and have a high reproducibility. However, slight variation in percentage of mortality can be observed, as the nematode is an in vivo model. To minimize those variations, worms and Bcc strains should be always maintained in the same conditions, using the same incubation times and temperatures.
- The principal concern relies on nematode’s larval stage. Only worms at larval stage 4 should be used. This is because worms at larval stage L3 or L2 can be more susceptible to pathogens, while older worms carry eggs, which can hamper the counting process.
- Timing is central. Please plan your experiment so that the experiment could be done using plates and animals that were prepared at the same time.
- Bcc strains can display different VR in the two assays. In our study we found a high variation in pathogenicity: strain B. metallica LMG 24068 and B. stabilis LMG 14294 had a maximum score in both assays, while B. cepacia LMG 1222 had VR = 3 in FKA and VR = 1 in SKA. B. multivorans LMG 13010 instead showed no pathogenicity at all VR = 0 in both assays.
Recipes
- LB broth (1 L)
10 g NaCl
10 g tryptone
5 g yeast extract
Add H2O to 1 L
Sterilize by autoclaving. - NGM agar medium (1L)
2.5 g peptone
17 g European agar
2.9 g NaCl
Add H2O to 1 L
Sterilize by autoclaving.
After solution cools down, add 1 ml autoclaved/sterile 1 M MgSO4, 1 ml autoclaved/sterile, 1 M CaCl2, 25 ml autoclaved/sterile 1 M KPO4, 1 ml cholesterol 5 mg/ml dissolved in 100% ethanol. - PGS agar medium (1 L)
12 g peptone
12 g glucose
27.25 g sorbitol
17 g European agar
2.9 g NaCl
Add H2O to 1 L
Sterilize by autoclaving.
After solution cools down, add 1 ml autoclaved/sterile 1 M MgSO4, 1 ml autoclaved/sterile, 1 M CaCl2, 25 ml autoclaved/sterile 1 M KPO4, 1 ml cholesterol 5 mg/ml dissolved in 100% ethanol. - M9 buffer (1 L)
3 g KH2PO4
6 g Na2HPO4
5 g NaCl
Add H2O to 1 L
Sterilize by autoclaving.
After solution cools down, add 1 ml autoclaved/sterile 1 M MgSO4.
Acknowledgments
Nematode strains used in this work were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). Burkholderia cepacia complex strains, used in this work, were kindly provided by Prof. Peter Vandamme, University of Gent, Belgium. This work was supported by the EU-KBBE 2012-2016 project PharmaSea, grant No. 312184.
References
- Cardona, S. T., Wopperer, J., Eberl, L. and Valvano, M. A. (2005). Diverse pathogenicity of Burkholderia cepacia complex strains in the Caenorhabditis elegans host model. FEMS Microbiol Lett 250(1): 97-104.
- Kothe, M., Antl, M., Huber, B., Stoecker, K., Ebrecht, D., Steinmetz, I. and Eberl, L. (2003). Killing of Caenorhabditis elegans by Burkholderia cepacia is controlled by the cep quorum-sensing system. Cell Microbiol 5(5): 343-351.
- Stiernagle, T. (2006). Maintenance of C. elegans. WormBook.
- Tedesco, P., Visone, M., Parrilli, E., Tutino, M. L., Perrin, E., Maida, I., Fani, R., Ballestriero, F., Santos, R., Pinilla, C., Di Schiavi, E., Tegos, G. and de Pascale, D. (2015). Investigating the role of the host multidrug resistance associated protein transporter family in Burkholderia cepacia complex pathogenicity using a Caenorhabditis elegans infection model. PLoS One 10(11): e0142883.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Tedesco, P., Di Schiavi, E., Palma Esposito, F. and de Pascale, D. (2016). Evaluation of Burkholderia cepacia Complex Bacteria Pathogenicity Using Caenorhabditis elegans. Bio-protocol 6(20): e1964. DOI: 10.21769/BioProtoc.1964.
- Tedesco, P., Visone, M., Parrilli, E., Tutino, M. L., Perrin, E., Maida, I., Fani, R., Ballestriero, F., Santos, R., Pinilla, C., Di Schiavi, E., Tegos, G. and de Pascale, D. (2015). Investigating the role of the host multidrug resistance associated protein transporter family in Burkholderia cepacia complex pathogenicity using a Caenorhabditis elegans infection model. PLoS One 10(11): e0142883.
Category
Neuroscience > Behavioral neuroscience > Animal model > Other
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












