- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Paper Roll Culture and Assessment of Maize Root Parameters
(*contributed equally to this work) Published: Vol 6, Iss 18, Sep 20, 2016 DOI: 10.21769/BioProtoc.1926 Views: 13735
Reviewed by: Tie LiuSriema L. WalawageArsalan Daudi

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Modified Pseudo-Schiff Propidium Iodide for Staining the Shoot Apical Meristem in Arabidopsis
Ruiqi Li [...] Ligeng Ma
May 5, 2023 2269 Views

A Novel Imaging Protocol for Investigating Arabidopsis thaliana Siliques and Seeds Using X-rays
Brylie A. Ritchie [...] Ansul Lokdarshi
Oct 5, 2023 2183 Views

Direct Plant Regeneration From Immature Male Inflorescence of Banana (Musa spp.)
Pradeep Chand Deo
Oct 20, 2025 1426 Views
Abstract
Selection for genotypes with a vigorous root system could enhance the adaptation of maize under water and nutrient deficit soils. Although extensive genetic variation for root architecture has been reported (Kumar et al., 2012; Abdel-Ghani et al., 2014; Kumar et al., 2014; Pace et al., 2015), root traits have been seldom considered as selection criteria to improve yield in maize, mainly because characterization of root morphology in the field is laborious, inaccurate and time consuming (Tuberosa and Salvi, 2007). Characterization of root traits under hydroponic conditions in this case has the advantage of screening a high number of genotypes in a small space (in a growth chamber) within a short period of time (2-3 weeks). Thus, it saves the time and effort required for screening maize genotypes with vigorous root systems and might be helpful to monitor root development at different growth stages.
Materials and Reagents
- Regular weight (brown) germination paper 48.5” x 36.5”, custom-sized to 12” x 24” (Anchor Paper Company, catalog number: SD3836S )
- Small kitchen wire mesh strainer/sieve with handle (20 cm diameter)
- Small plastic cups, measuring boats (FisherbrandTM Hexagonal Polystyrene Weighing dishes) (Thermo Fisher Scientific, Fisher Scientific, catalog number: 02-202-101 ), or double-faced filter paper for drying seed- should be the same number as the number of entries
- Waterproof pencil art grip aquarelle black (Faber-Castell, catalog number: 114299 ) and permanent marker (Sharpie®, Fine Point Permanent Marker, black)
Note: Black marker works better as other colors fade faster. - Plastic tags (5” x 5/8”) (International Greenhouse, catalog number: CN-1000 ) for labeling (optional if rolls are labeled)
- Rubber bands (OfficeMax Extra Long Rubber bands, or any other brand)
- Glassine bags (Seedburo S411 shoot bags, treated, 4” x 2-1/2” x 11”)
- Personal protective items: latex gloves, lab coat, closed shoes, mask goggles
- Maize seeds: Genotypes used in this protocol are pure lines obtained from the North Central Regional Plant Introduction Station in Ames, Iowa (Abdel-Ghani et al., 2013).
Notes:- Seeds should be multiplied under the same conditions to avoid differences due to the environment on the seed size.
- Seeds should display high germination percentages to keep a similar number of biological replications within experimental units.
- Seeds should be multiplied under the same conditions to avoid differences due to the environment on the seed size.
- Chlorox® solution (6% sodium hypochlorite), household bleach (USA)
- Deionized sterile distilled water (ddH2O)
- Potassium nitrate (KNO3) (Thermo Fisher Scientific, Fisher Scientific, catalog number: P-263-500 )
- Calcium nitrate [Ca(NO3)2] (Thermo Fisher Scientific, Fisher Scientific, catalog number: C109-3 )
- Monopotassium phosphate or potassium phosphate monobasic (KH2PO4) (Thermo Fisher Scientific, Fisher Scientific, catalog number: BP-362-500 )
- Magnesium sulfate (Thermo Fisher Scientific, Fisher Scientific, catalog number: M65-500 )
- Iron from iron chelate [Fe-EDTA, (Sigma-Aldrich, catalog number: E6760-100G ), Fe-DTPA, or Fe-EDDHA]
- Monocalcium phosphate or calcium phosphate monobasic [Ca(H2PO4)2] (MP Biomedicals, catalog number: 193803 )
- Calcium sulfate dihydrate (CaSO4·2H2O) (MP Biomedicals, catalog number: 191414 )
- Potassium sulfate (K2SO4) (Sigma-Aldrich, catalog number: P0772-1kg )
- Boric acid (Thermo Fisher Scientific, Fisher Scientific, catalog number: BP168-1 )
- Manganese chloride-4 hydrate (MnCl2·4H2O) (Sigma-Aldrich, catalog number: 221279-100g )
- Zinc sulfate-7 hydrate (ZnSO4·7H2O) (Sigma-Aldrich, catalog number: Z0251-100G )
- Copper sulfate-5 hydrate (Thermo Fisher Scientific, Fisher Science, catalog number: S25287A )
- Molybdic acid (H2MoO4) (Sigma-Aldrich, catalog number: 232084-100G )
- Hoagland’s nutrient solution
- High N (15 mM NO3-) Hoagland’s solution (see Recipes)
- Low N (1.5 mM NO3-) Hoagland’s solution (see Recipes)
- Micronutrient stock solution (1 L) (see Recipes)
- High N (15 mM NO3-) Hoagland’s solution (see Recipes)
- 30% ethanol (C2H6O) (commercial grade from any brand) (see Recipes)
- 2.5 g/L Fungicide solution Captan® (Bonide Products Inc.) (see Recipes)
Equipment
- 2 L capacity beakers (each beaker holds 8-10 paper rolls) (Coring, Pyrex® Griffin Beakers, catalog number: 10000-2L )
- 50 ml capacity beakers (Pyrex®, Griffin Beakers), for sterilizing and washing seeds (Sigma-Aldrich, catalog number: CLS100050 )
- Plant growth chamber (Conviron, model: PGC FLEX )
- Cold room (Rheem Puffer Hubbard environmental chamber) or refrigerator
- Autoclave (PRIMUS Sterilizer , model: PSS5 )
- Sensitive balance (Ohaus, model: AdventurerTM AR0640 )
Note: This product has been discontinued. - Flatbed scanner (Epson, model: Expression 10000 XL , or any other brand)
- Computer with flash drive and Windows operating system
- Fisher ScientificTM IsotempTM general purpose heating and drying oven (Thermo Fisher Scientific, Fisher Scientific, model: 15-103-0503) or any constant temperature oven/dryer
- Ruler or yardstick (Acme Westcott 15728 36” Aluminum Yard Stick)
Software
- WinRhizo (Regent Instruments, model: WinRhizo Pro 2009) or ARIA (Automatic Root Image Analysis) (Pace et al., 2014)
Procedure
This experiment was designed to test the performance of maize seedlings under contrasting level of nitrogen (N) levels (Abdel-Ghani et al., 2013). The seedlings should be exposed to Hoagland’s nutrient solution with high N (HN) and low N (LN) (Hershey, 1994). Nitrogen in Hoagland’s solution with HN contains 15 mM of NO3-, whereas the concentration of N in LN Hoagland’s solution is 1.5 mM (10% NO3-) (Abdel-Ghani et al., 2013; Abdel-Ghani et al., 2015). Other macro- and micro-elements should be constant in both nutrient treatments. All steps regarding paper roll preparation and culture are as follows (Also see Figure 1; steps A to D).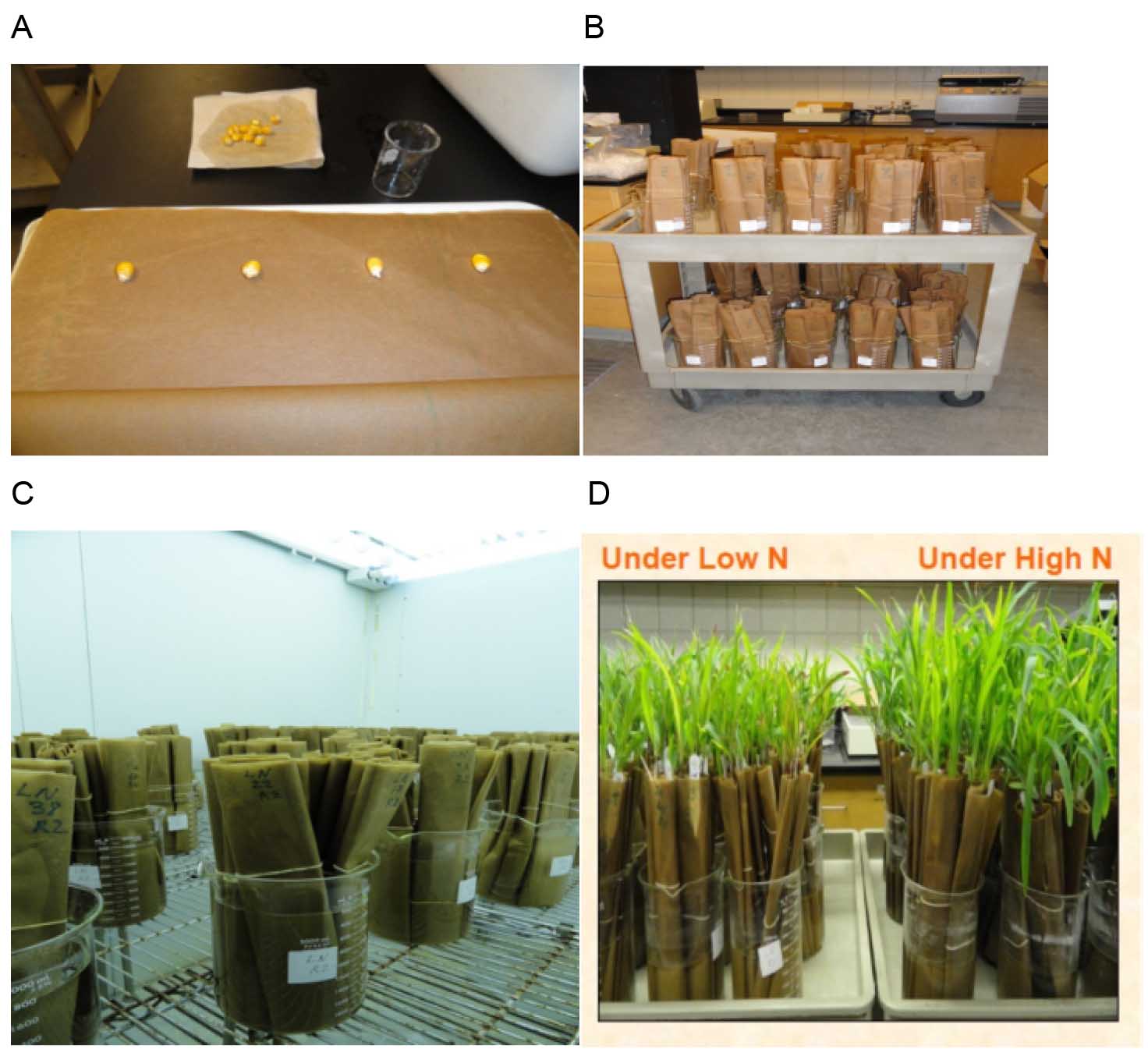
Figure 1. Summary of steps for paper rolls preparation and culture. A. Kernels are surface sterilized with 6% sodium hypochlorite, washed with distilled water and dried out. Four sterilized maize kernels of similar size are placed on a double layer of brown filter papers pre-moistened with fungicide solution Captan®. B. Rolled germination papers are kept in 2 L glass beakers containing Hoagland’s nutrient solution with high nitrogen (HN) and low nitrogen (LN). C. Rolls should be kept for 14 days in a controlled growth chamber. D. Seedling after 14 days of incubation in the controlled growth chamber, under LN and HN treatments.
- Maize kernels sterilization
- Kernels are surface sterilized in 20 ml beakers with Chlorox® solution (6% sodium hypochlorite) for 15 min at room temperature. Chlorox should cover the kernels in the beaker and beakers should be manually shaken for 3-4 times.
- Chlorox should be drained out first, and then the seeds should be washed with ddH2O three times. Small sterilized sieve (20 cm in diameter) can be used to drain out water after each wash.
- After washing, kernels should be kept on a double-faced brown filter paper and left for 10 min until the seeds are dry. Small plastic cups or measuring boats can also be used to dry seeds.
- Kernels are surface sterilized in 20 ml beakers with Chlorox® solution (6% sodium hypochlorite) for 15 min at room temperature. Chlorox should cover the kernels in the beaker and beakers should be manually shaken for 3-4 times.
- Growing seeds in paper rolls
- Brown germination paper should be cut down into 20 x 20 cm sheets and pre-moisturized with fungicide solution Captan® (2.5 g/L) to eliminate the possibility of any fungi development during seedling development. The brown paper should be moistened with fungicide solution by soaking the paper in the solution. Excess fungicide solution should be removed by pressing on the soaked papers by hand. The paper rolls should be labeled either with a permanent (water proof) pencil by writing directly on brown sheets and/or by attaching labels with each roll.
- Four sterilized maize kernels of similar size are placed 4 cm away from the top edge of a double layer of filter papers. Kernels are placed 4 cm apart and leaving 4 cm from left and right edges, covered with another filter paper, then wrapped into rolls, about 5 cm thick. The roll should be kept secure with a rubber band. Two-L capacity glass beakers containing autoclaved Hoagland’s nutrient solution with HN or LN should be filled to one half (about 400 ml), and consequently brown paper rolls should be placed vertically in the beakers, making sure that the seeds are on top and not submerged in the solution. About one half of the length of the rolls should be emerged in the solution. Eight to ten rolls could be placed per beaker. All steps were illustrated in Figure 1A-D.
- Brown germination paper should be cut down into 20 x 20 cm sheets and pre-moisturized with fungicide solution Captan® (2.5 g/L) to eliminate the possibility of any fungi development during seedling development. The brown paper should be moistened with fungicide solution by soaking the paper in the solution. Excess fungicide solution should be removed by pressing on the soaked papers by hand. The paper rolls should be labeled either with a permanent (water proof) pencil by writing directly on brown sheets and/or by attaching labels with each roll.
- Growing conditions
Rolls should be kept in a controlled growth chamber under the following conditions (Figure 1D): a photoperiod of 16/8 h (light/darkness) at 25/22 °C with photosynthetically active radiation of 200 µmol photons m-2 s-1. The relative humidity in the growth chamber should be maintained at 65%. Nutrient solution should be daily added to maintain the solution level in the beakers at 400 ml during the experiment. Seedlings should be kept 14 days in the growth chamber. Thereafter, maize root architecture related traits could be recorded either manually or using image analysis software. - Recording maize root architecture
After 14 days of incubation of maize kernels grown under HN and LN levels, the nutrient solution should be removed and replaced with about 400 ml of 30% ethanol and the samples should be stored in a cold room, only to be taken out for measuring, scanning, and drying. This is done to prevent further growth in order to preserve the roots and to record root data at the same time point. However, this step is not necessary if all roots can be scanned in one day. For measuring root traits using software, scan the roots and save the images using a flatbed scanner, as much as possible, make sure that roots do not overlap for ease in measurement. Out of the 4 seedlings per entry, 3 that look similar would be measured. Scanning should be done before drying the roots.- Manually recorded traits: the root of individual seedlings should be separated into three parts by a blade, namely, primary root, seminal roots and crown roots (Figure 2). Maize root traits could be recorded by a ruler or gravimetrically. The lengths of primary root, seminal roots and crown roots can be recorded by a ruler. Seminal root number and crown root number can be recorded by counting up the rising roots. Fresh weight of shoots and roots could also be recorded using sensitive balance. Fresh roots are put in glassine paper bags (10 x 20 cm) and oven dried at 80 °C for at least 48 h. Dry weight measurements can be taken after drying using a sensitive balance.
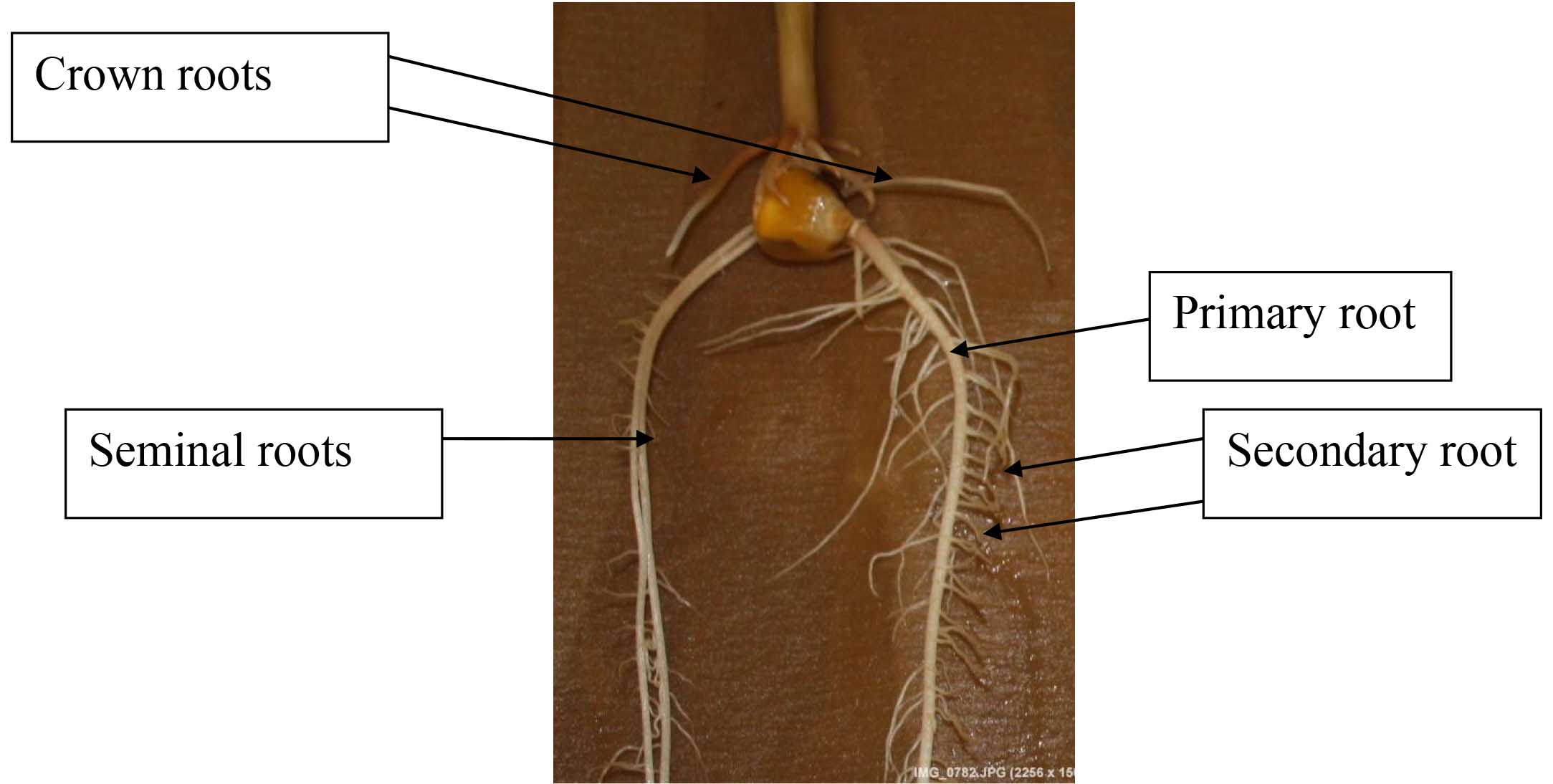
Figure 2. Maize root system: embryonic roots (primary roots and seminal roots) and postembryonic roots (shoot born crown roots and lateral roots). Crown roots are responsible for the major part of the water and nutrient uptake. All these roots are usually formed below the soil. - Traits recorded by WinRhizo program: total number of root tips, forks, and crossings, total root length, root surface area, root volume and root average diameter could be measured using image analysis software. The software cannot distinguish primary, seminal, crown, or lateral roots by itself. As mentioned in the previous step, the roots have to be divided into primary, seminal, and crown roots; then, specific root measurements can be done. Steps of root imaging analysis using WinRhizo software are presented in Figures 3-17.
- Turn the scanner power on, open WinRhizo program and select the scanner that will be used (should be highlighted), then click “Select” (Figure 3).
Figure 3. Scanner (image source) selection in WinRhizo - The title window for WinRhizo will open. Click “Ok” (Figure 4).
Figure 4. WinRhizo Startup page. Click “Ok” to continue. - Scanning and saving root images
- Place up to 3 roots on the scanner. The root system can be entirely scanned or separate scans of dissected root parts could be performed in the case of a very dense and compacted root system. Make sure that the roots from one plant are not intertwined with those of other plants, as this affects the analysis.
- From the main tab, click “Image”, then click “Acquire Image” (Figure 5).
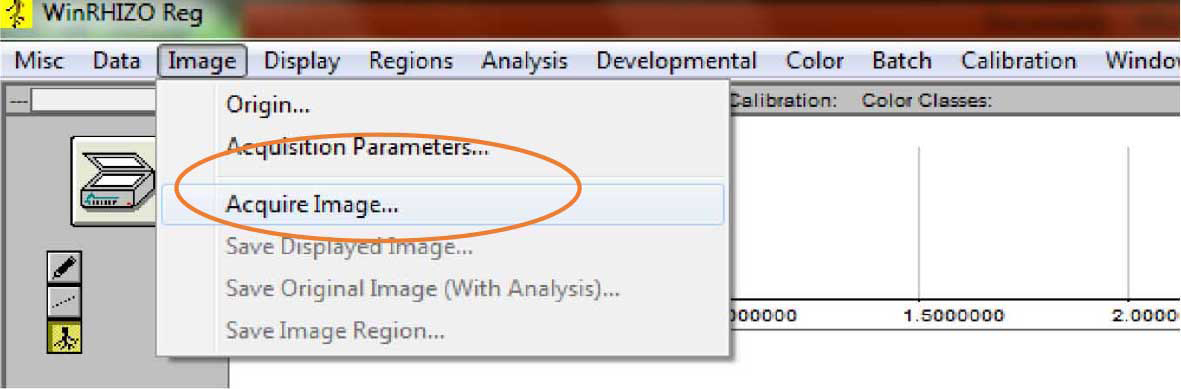
Figure 5. Image acquisition in WinRhizo. After clicking on “Image>Acquire Image,” scanning of roots commences. - The root images will show on the screen once scanning is done. Check the images (Figure 6).
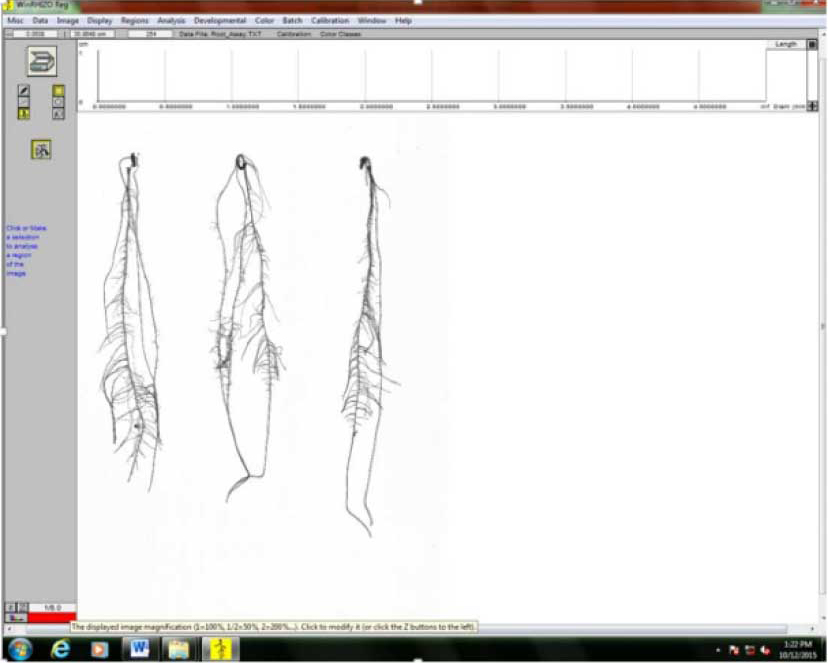
Figure 6. Scanned roots image preview from WinRhizo software - Save the scanned image by clicking “Image,” then “Save Displayed Image”.
- Analyzing scanned root images
- Make a new file where the root parameter data will be stored. From the main tab, click “Data”, then click “New File.” If you have previously started the analysis, then select “Open file” (Figure 7).
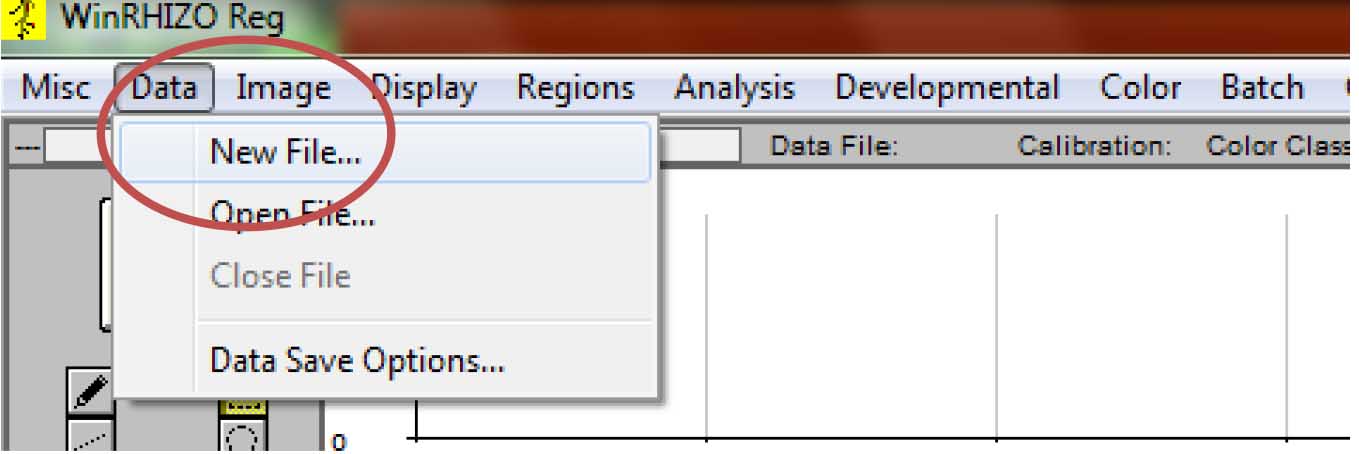
Figure 7. Creating/opening a file where the root parameter data will be saved. If analysis is done for the first time in a particular experiment, select “Image>New File…” If analysis for the same experiment has started. Select “Image>Open File…”. - Select the location where the file will be saved. It is advisable to save the file in the folder where the images are. Type the desired file name, then click “Save.” The data file will be in text format (.txt) (Figure 8).
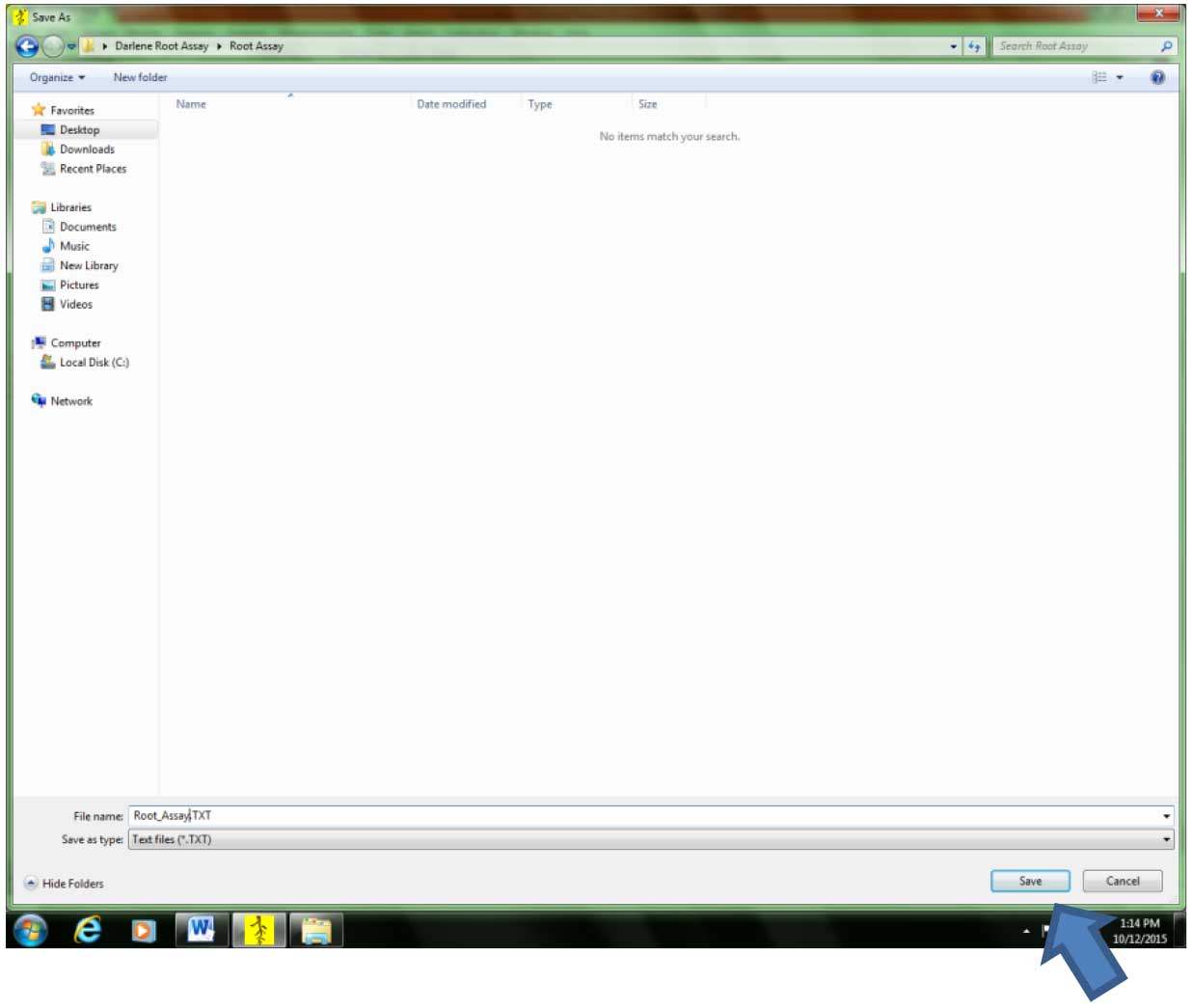
Figure 8. Selecting the directory/folder where the data file will be saved - To get the scanned images, click “Image”, the “Origin.” A new window (“Image Origin”) will appear. Select Disk (Figure 9).
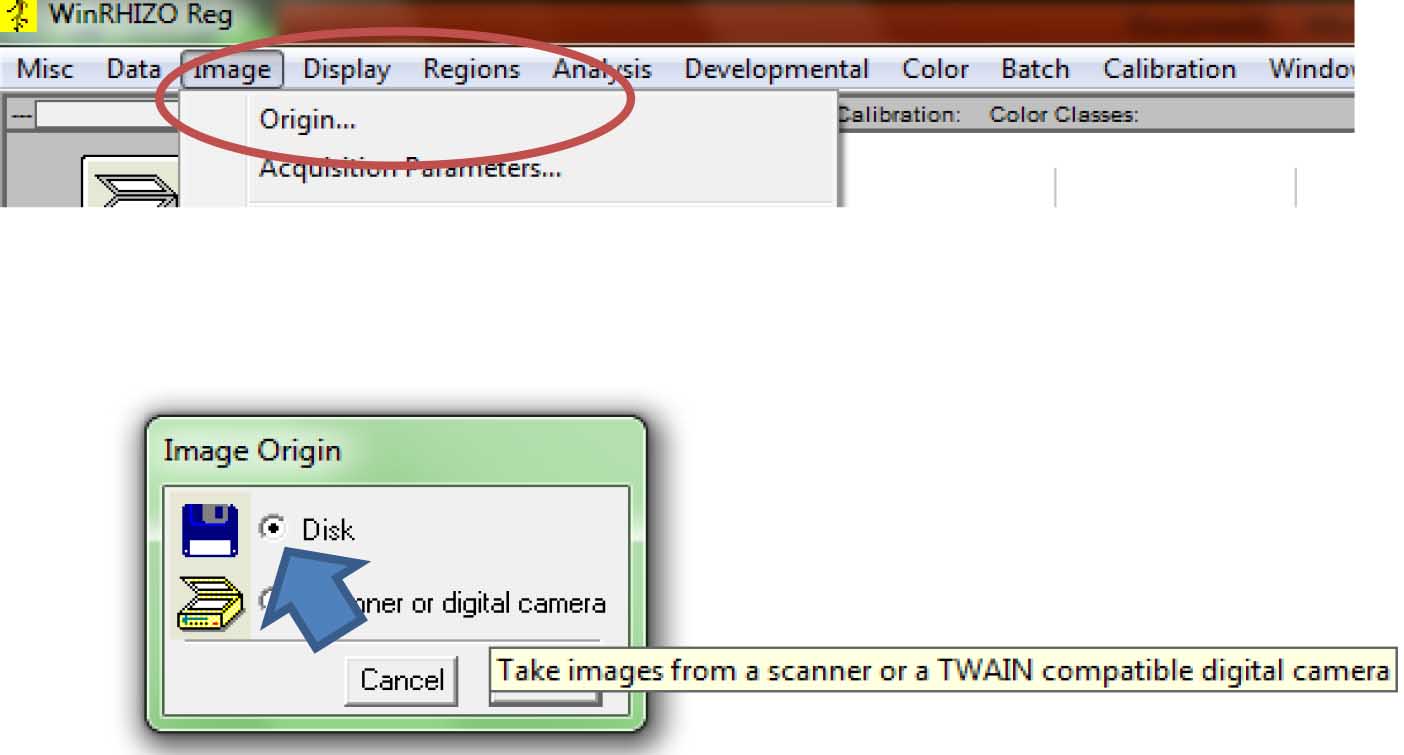
Figure 9. Selecting the source of the root images for analysis (Click on “Image>Origin”) - To acquire the saved images, click on the floppy disk icon on the upper left side of the screen (Figure 10).
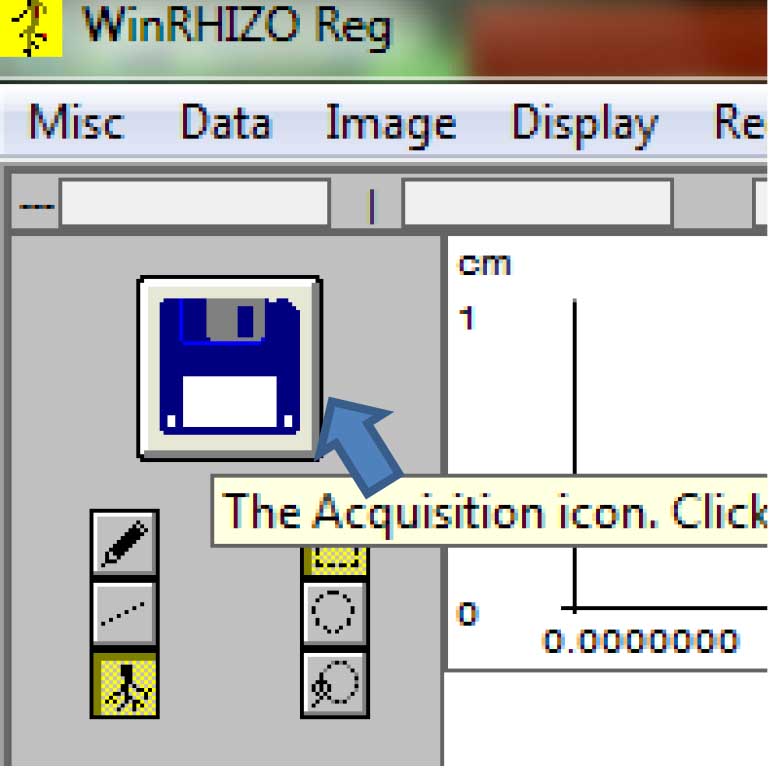
Figure 10. Acquiring the previously scanned images for analysis. Click on the floppy disk icon. - Click on the image to be analyzed, then click “Open” (Figure 11).
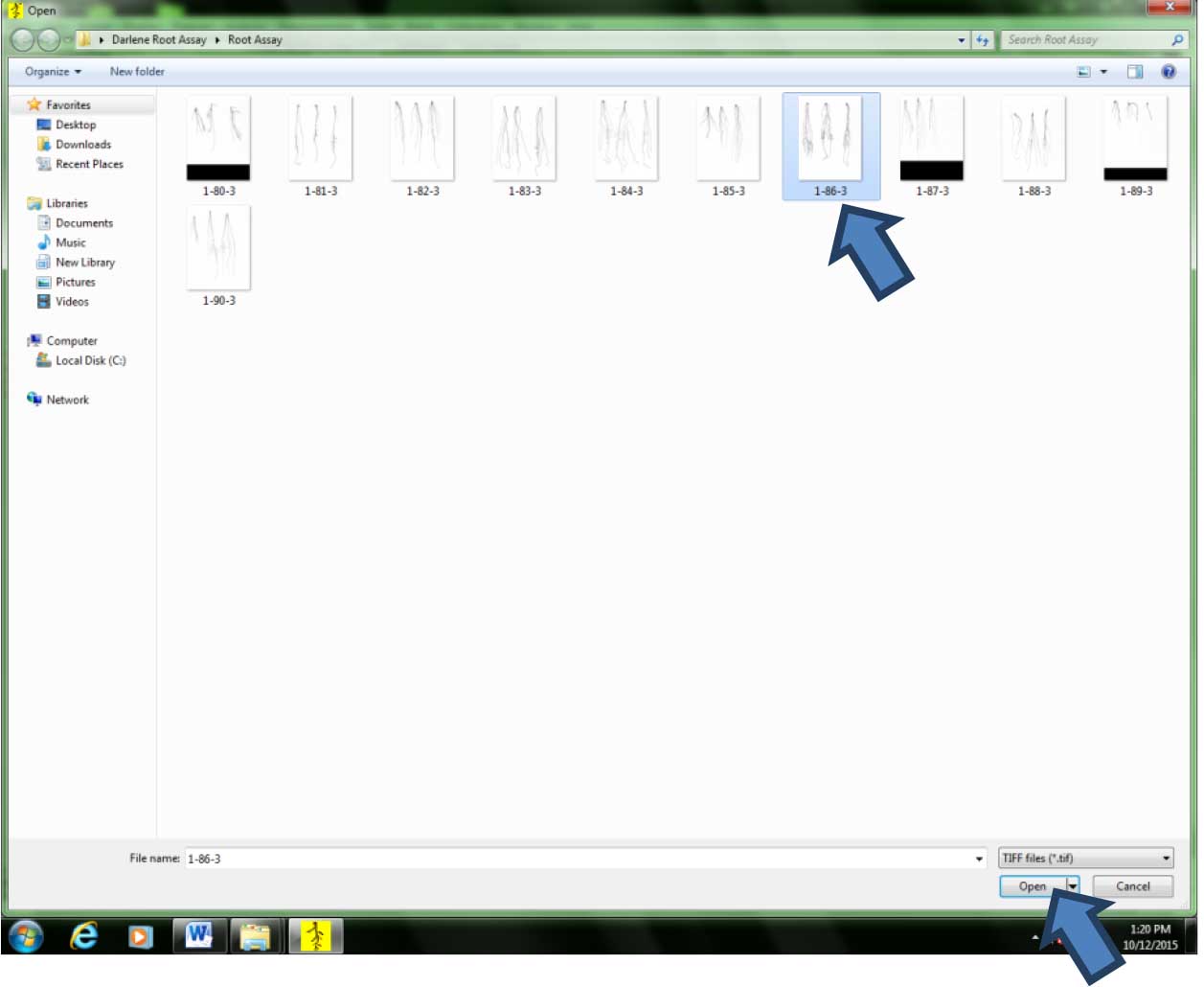
Figure 11. Selecting the image to be analyzed by clicking on the file thumbnail - Zoom the image out so that the whole image can be seen on the screen. In this example, the image was zoomed out to 1/8th of its original size (Figure 12).
1) Original size (Figure 12A)
2) Zoomed-out image (Figure 12B)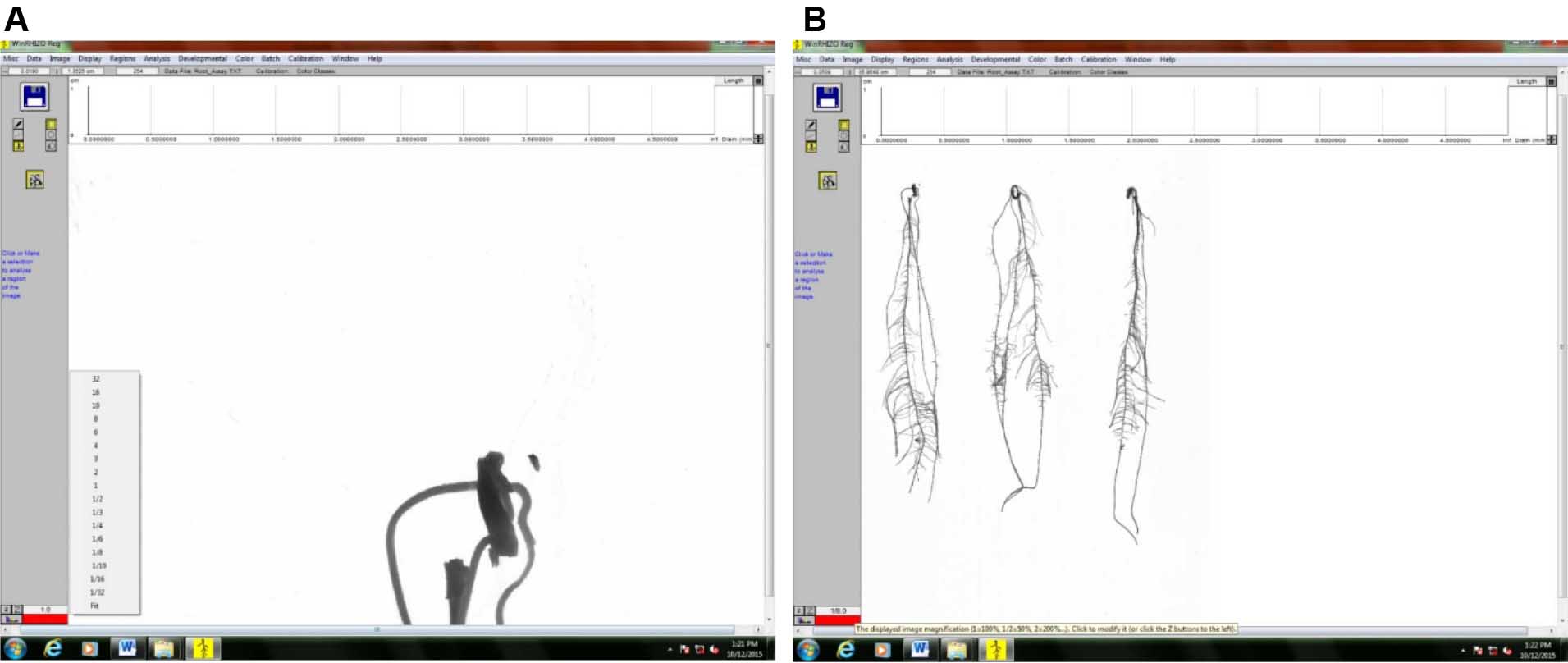
Figure 12. Image of scanned roots for analysis. A. Original size; B. Image zoomed out to 1/8th of its size to show all roots. - Select the root to be analyzed by first clicking either: 1). Rectangular selection or; 2). Free form selection for closely-spaced roots (Figure 13).
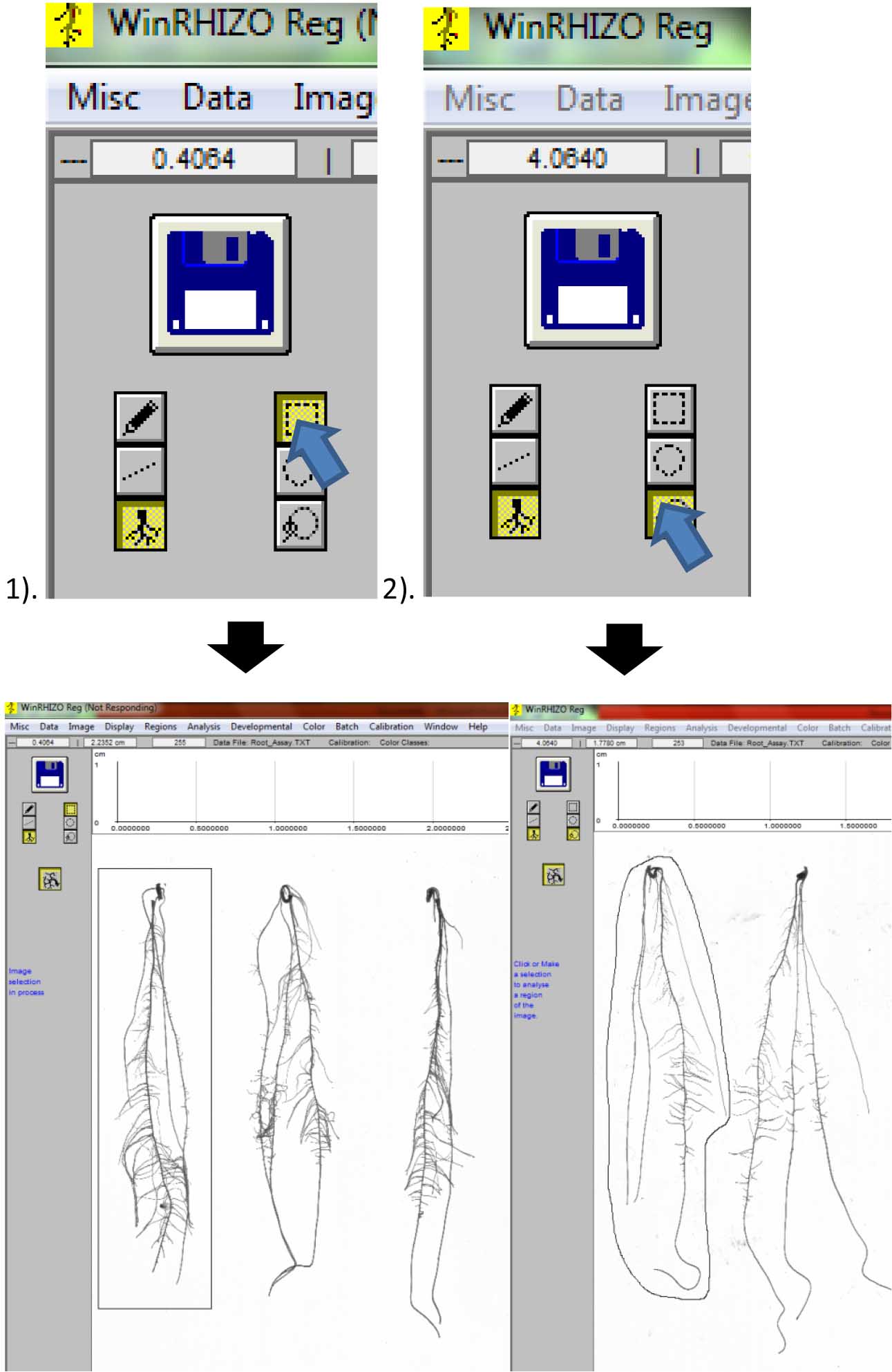
Figure 13. Selecting individual roots for analysis. 1). Rectangular selection; 2). Free form selection. - A window will appear. Label the root beside “Identification” and write your name as “Operator” Then click “Ok” (Figure 14).
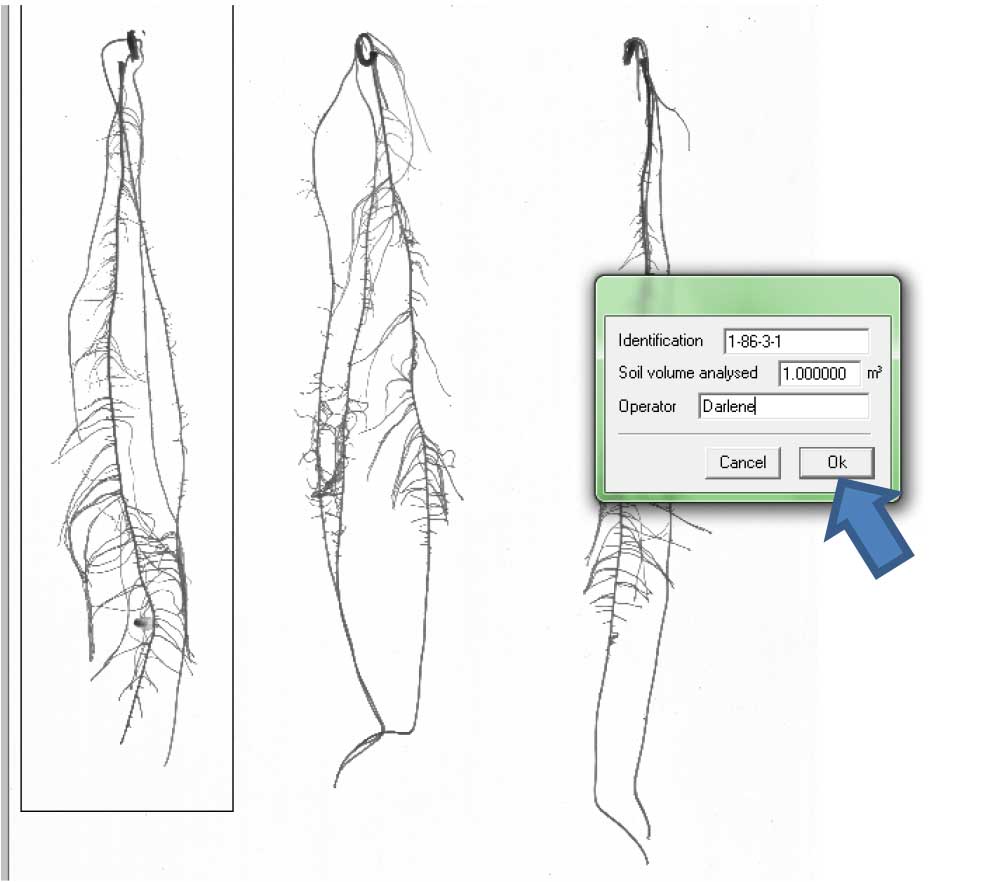
Figure 14. Labeling the individual roots - Repeat steps D2 vii-viii until all the roots are analyzed (Figure 15).
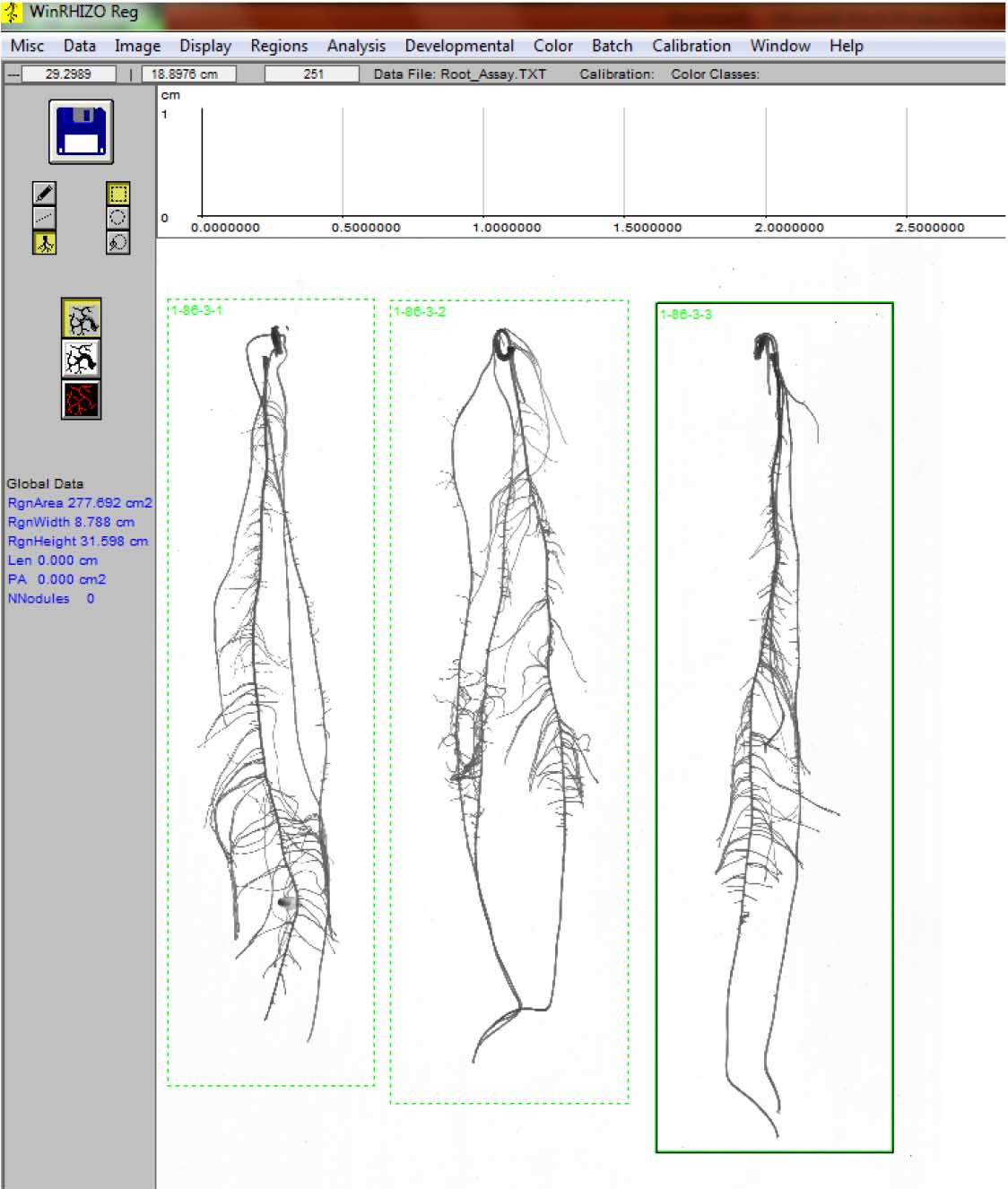
Figure 15. End of root image analysis, indicated by green outlines and labels at the upper left side for each root - The output file looks like this: It is in text (*.txt) format; open the file using MS Excel and save as Excel spreadsheet to be able to organize (sort, filter) and make calculations (e.g., average) with the data. Root morphological data from individual plants can be found in the following columns (Figure 16):
Length (cm) (Column 16/P) – Total root length (cm)
ProjArea (cm2) (Column 18/R) – Total root projected area (cm2)
SurfArea (cm2) (Column 20/T) – Total root surface area (cm2)
AvgDiam (mm) (Column 22/V) – Average root diameter (mm)
LenPerVol (cm/m3) (Column 24/X) – Total root length per cubic meter of soil (cm/m3)
RootVolume (cm3) (Column 26/Z) – Total root volume
Tips (Column 29/AC) – Number of tips
Forks (Column 30/AD) – Number of forks
Crossings (Column 31/AE) – Number of crossings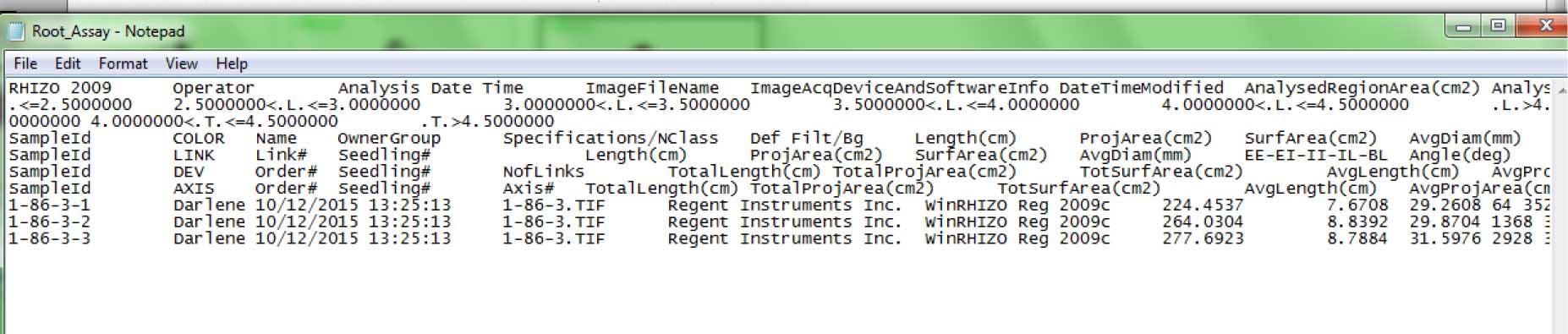
Figure 16. Sample output file (in *.txt format) from root imaging analysis using WinRhizo software (step D2x)
- Turn the scanner power on, open WinRhizo program and select the scanner that will be used (should be highlighted), then click “Select” (Figure 3).
- Traits recorded by ARIA program: steps of root imaging analysis using ARIA program are presented in Figure 17. The measurements are done by the software all at once. After loading the images and clicking on the primary root on the image (the same can be done for other roots), the software will start measuring all traits at the same time. The following traits and their corresponding description can be measured using ARIA (Pace et al., 2014).
- Total root length (TRL) – Cumulative length of all the roots in centimeters
- Primary root length (PRL) – Length of the primary root in centimeters
- Secondary root length (SEL) – Cumulative length of all secondary roots in centimeters
- Center of mass (COM) – Center of gravity of the root.
- Center of point (COP) – Absolute center of the root regardless of root length.
- Center of mass (Top) (CMT) – Center of gravity of the top 1/3 of the root (Top).
- Center of mass (Mid) (CMM) – Center of gravity of the middle 1/3 root (Middle).
- Center of mass (Bottom) (CMB) – Center of gravity of the bottom 1/3 root (Bottom).
- Center of point (Top) (CPT) – Absolute center of the root regardless of root length (Top).
- Center of point (Mid) (CPM) – Absolute center of the root regardless of root length (Middle).
- Center of point (Bottom) (CPB) – Absolute center of the root regardless of root length (Bottom).
- Maximum number of roots (MNR) – The 84th percentile value of the sum of every row.
- Perimeter (PER) – Total number of network pixels connected to a background pixel.
- Depth (DEP) – The maximum vertical distance reached by the root system.
- Width (WID) – The maximum horizontal width of the whole RSA.
- Width/Depth ratio (WDR) – The ratio of the maximum width to depth.
- Median (MED) – The median number of roots at all Y-location.
- Total number of roots (TNR) – Total number of roots.
- Convex area (CVA) – The area of the convex hull that encloses the entire root image
- Network area (NWA) – The number of pixels that are connected in the skeletonized image
- Solidity (SOL) – The fraction equal to the network area divided by the convex area
- Bushiness (BSH) – The ratio of the maximum to the median number of roots.
- Length distribution (LED) – The ratio of TRL in the upper one-third of the root to the TRL.
- Diameter (DIA) – Diameter of the primary root.
- Volume (VOL) – Volume of the primary root
- Surface area (SUA) – Surface area of the primary root.
- SRL – Total root length divided by root system volume
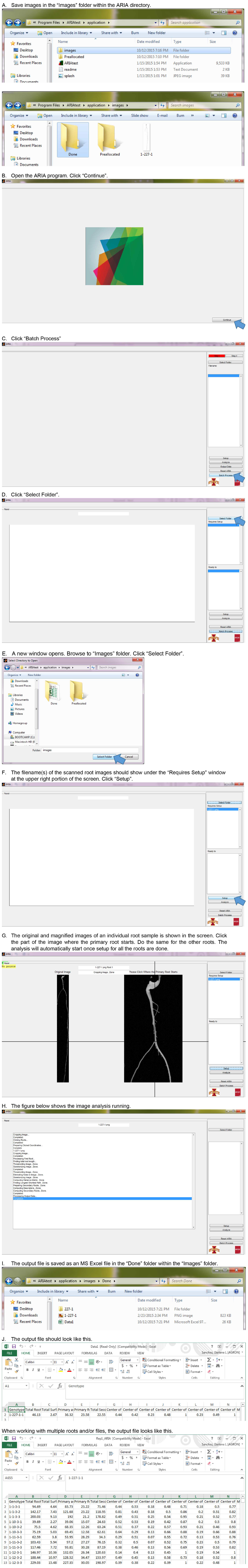
Figure 17. Root image analysis using ARIA software (step D3).
- Total root length (TRL) – Cumulative length of all the roots in centimeters
- Manually recorded traits: the root of individual seedlings should be separated into three parts by a blade, namely, primary root, seminal roots and crown roots (Figure 2). Maize root traits could be recorded by a ruler or gravimetrically. The lengths of primary root, seminal roots and crown roots can be recorded by a ruler. Seminal root number and crown root number can be recorded by counting up the rising roots. Fresh weight of shoots and roots could also be recorded using sensitive balance. Fresh roots are put in glassine paper bags (10 x 20 cm) and oven dried at 80 °C for at least 48 h. Dry weight measurements can be taken after drying using a sensitive balance.
Representative data
Representative data showing seedling growth of genotypes PHZ51, B73 and Mo17 under LN and HN levels are shown in Figure 18. Maize genotypes responded to N deficiency by increasing the root:shoot (R:S) ratio. Lines presented in Figure 18 displayed a higher R:S ratio under LN as compared with HN treatment. To absorb sufficient amount of N under LN, maize plants adapt to N starvation by increasing the root volume and decreasing the aerial vegetative growth.
Figure 18. Performance of three maize genotypes (PHZ51, B73 and Mo17) under low and high nitrogen levels.
Recipes
- Hoagland’s nutrient solution
Note: This is made essentially according to Hoagland and Arnon (1950). This solution is used to culture plants in hydroponic medium.- High N (15 mM NO3-) Hoagland’s solution
5 ml of 1 M (M = Molar) potassium nitrate
5 ml of 1 M calcium nitrate
1 ml of 1 M monopotassium phosphate
2 ml of 1 M magnesium sulfate
1 ml of micronutrient stock solution (see Recipe 1c below)
5 ml of 1,000 mg/L iron from iron chelate (Fe-EDTA, Fe-DTPA, or Fe-EDDHA)
Add ddH2O to complete the volume to 1 L - Low N (1.5 mM NO3-) Hoagland’s solution
10 ml of 0.05 M monocalcium phosphate
200 ml of 0.01 M calcium sulfate dihydrate
5 ml of 0.5 M potassium sulfate
2 ml of 1 M magnesium sulfate
1 ml of micronutrient stock solution (see Recipe 1c below)
5 ml of iron chelate stock solution as for HN
Add ddH2O to complete the volume to 1 L - Micronutrient stock solution (1 L)
2.86 g boric acid
1.81 g manganese chloride-4 hydrate
0.22 g zinc sulfate-7 hydrate
0.08 g copper sulfate-5 hydrate
0.02 g 85% molybdic acid
Add autoclaved ddH2O to complete the volume to 1 L
- High N (15 mM NO3-) Hoagland’s solution
- 30% ethanol (1 L)
Add gradually 300 ml ethanol in 500 ml of ddH2O by stirring
Add ddH2O until final volume is 1,000 ml
Store at 4 °C - 2.5 g/L Fungicide solution Captan®
Dissovle 25 g of Captan® in 500 ml of ddH2O by stirring
Add ddH2O until final volume is 1,000 ml
Store at room temperature
Acknowledgments
This protocol was developed while Dr. Adel Abdel-Ghani was a visiting Fulbright Postdoctoral Fellow and during the sabbatical leave granted from Mu’tah University, Jordan during the academic year 2011-2012 at Iowa State University (ISU), Ames, USA. Authors would also like to thank USDA’s National Institute of Food and Agriculture (project number: IOW05180) and RF Baker Center for Plant Breeding for funding this work.
References
- Abdel-Ghani, A. H., Kumar, B., Pace, J., Jansen, C., Gonzalez-Portilla, P. J., Reyes-Matamoros, J., San Martin, J. P., Lee, M. and Lubberstedt, T. (2015). Association analysis of genes involved in maize (Zea mays L.) root development with seedling and agronomic traits under contrasting nitrogen levels. Plant Mol Biol 88(1-2): 133-147.
- Abdel-Ghani, A. H., Kumar, B., Reyes-Matamoros, J., Gonzalez-Portilla, P. J., Jansen, C., San Martin, J. P., Lee, M. and Lubberstedt, T. (2013). Genotypic variation and relationships between seedling and adult plant traits in maize (Zea mays L.) inbred lines grown under contrasting nitrogen levels. Euphytica 189(1): 123-133.
- Arana, M. V., Sanchez-Lamas, M., Strasser, B., Ibarra, S. E., Cerdan, P. D., Botto, J. F. and Sanchez, R. A. (2014). Functional diversity of phytochrome family in the control of light and gibberellin-mediated germination in Arabidopsis. Plant Cell Environ 37(9): 2014-2023.
- Hoagland, D. R. and Arnon, D. I. (1950). The water-culture method for growing plants without soil. Calif Agric Exp Stn Circ 347:1-32.
- Kumar, B., Abdel-Ghani, A. H., Pace, J., Reyes-Matamoros, J., Hochholdinger, F. and Lubberstedt, T. (2014). Association analysis of single nucleotide polymorphisms in candidate genes with root traits in maize (Zea mays L.) seedlings. Plant Sci 224: 9-19.
- Kumar, B., Abdel-Ghani, A. H., Reyes-Matamoros, J., Hochholdinger, F. and Lubberstedt, T. (2012). Genotypic variation for root architecture traits in seedlings of maize (Zea mays L.) inbred lines. Plant Breeding 131(4): 465-478.
- Pace, J., Gardner, C., Romay, C., Ganapathysubramanian, B. and Lubberstedt, T. (2015). Genome-wide association analysis of seedling root development in maize (Zea mays L.). BMC Genomics 16: 47.
- Pace, J., Lee, N., Naik, H. S., Ganapathysubramanian, B. and Lubberstedt, T. (2014). Analysis of maize (Zea mays L.) seedling roots with the high-throughput image analysis tool ARIA (Automatic Root Image Analysis). PLoS One 9(9): e108255.
- Tressler, W. L. and Williams, T. (1937). Hydroponics solution used for daphnia culture. Science 86(2229): 273-274.
- Tuberosa, R. and Salvi, S. (2007). From QTLs to genes controlling root traits in maize. Scale and Complexity in Plant Systems Research: Gene-Plant-Crop Relations 21: 15-24.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Abdel-Ghani, A. H., Sanchez, D. L., Kumar, B. and Lubberstedt, T. (2016). Paper Roll Culture and Assessment of Maize Root Parameters. Bio-protocol 6(18): e1926. DOI: 10.21769/BioProtoc.1926.
Category
Plant Science > Plant developmental biology > Morphogenesis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










