- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Actin Retrograde Flow in Permeabilized Cells: Myosin-II Driven Centripetal Movement of Transverse Arcs
Published: Vol 6, Iss 5, Mar 5, 2016 DOI: 10.21769/BioProtoc.1743 Views: 9609
Reviewed by: Lin FangRalph BottcherAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Lipid-Mediated Sequential Recruitment of Proteins Via Dual SLIPT and Dual SLIPTNVOC in Live Cells
Kristina V. Bayer and Richard Wombacher
Nov 5, 2025 1600 Views

Monitoring of Sperm-Independent Calcium Oscillations in Immature Oocytes of Mice
Sae Horiike [...] Hidehiko Ogawa
Feb 5, 2026 65 Views

High Content In Vitro Survival Assay of Cortical Neurons
Paolo V. Fioretti [...] Manuela Basso
Feb 5, 2026 80 Views
Abstract
Numerous biological functions such as cytokinesis, changes in cell shape and cell migration require actomyosin-driven cellular contractility. However, the detailed mechanism of how contractile forces drive cellular processes are difficult to decipher due to the complexity of the intracellular environment. In particular, the mesoscopic description of the myosin II-dependent actin retrograde flow in cell lamellum is missing. Here, we describe a methodology for detergent extraction of cell, which preserves integrity of the actin cytoskeleton. This semi-in vitro cell model allows for the observation, using light microscopy, and quantification of changes in the actin cytoskeleton resulting from the activation of cellular contractility upon addition of ATP. This assay also allows for the evaluation of the effects of actin-associated proteins and other related factors in the modulation of the actin contractile activities. Here, we demonstrate the retrograde flow of a well-known actin-based structures- transverse arcs, which are myosin IIA-containing structures that emerge at the boundary between lamellipodium-lamellum and move centripetally in myosin II-dependent fashion.
Keywords: Actin fibersMaterials and Reagents
- 35-mm ibidi’s hydrophobic uncoated μ-dishes for cell culture (ibidi GmbH, catalog number: 80131 )
- 1 x 1 cm polydimethylsiloxane (PDMS) stamp containing microfeatures of circles (area, 1,800 μm2; center-to-center distance, 100 μm)
Note: For a detailed protocol on preparation of PDMS stamps and micro-contact printing see Théry and Piel (2009) and Tee et al., (2015). - NuncTM Cell Culture Treated Flasks with Filter Caps (Thermo Fisher Scientific, catalog number: 136196 )
- Human foreskin fibroblast (ATCC, catalog number: SCRC-1041 )
- Growth medium: Dulbecco’s modified Eagle’s medium (DMEM) high glucose (Thermo Fisher Scientific, catalog number: 11965-092 ), supplemented with
- TrypLETM Express Enzyme (Thermo Fisher Scientific, catalog number: 12604013 )
- PBS (1x), pH 7.4 (Thermo Fisher Scientific, catalog number: 10010-023 )
- Fibronectin (Merck Millipore Corporation, catalog number: 341635 )
- Imidazole (Sigma-Aldrich, catalog number: I15513 )
- KCl (First BASE Laboratories Sdn Bhd, catalog number: BIO-1300 )
- Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: M8266 )
- EDTA (First BASE Laboratories Sdn Bhd, catalog number: BIO-1050 )
- Ethylene glycol-bis (2-aminoethylether)-N, N, N′, N′-tetraacetic acid (EGTA) (Sigma-Aldrich, catalog number: E3889 )
- 2-Mercaptoethanol (Sigma-Aldrich, catalog number: M6250 )
- TritonTM X-100 (Sigma-Aldrich, catalog number: X100 )
- Polye (ethylene glycol) MW35,000 (PEG) (Sigma-Aldrich, catalog number: 81310 )
- Protease inhibitors cocktail (for use with mammalian cell and tissue extracts, DMSO solution) (Sigma-Aldrich, catalog number: P8340 )
- N, N-Dimethylformamide (DMF) (Sigma-Aldrich, catalog number: 227056 )
- Methanol (Thermo Fisher Scientific, catalog number: M/4000/17 )
- Sterile Milli-Q water
- Alexa Fluor® 488 Phalloidin (Thermo Fisher Scientific, catalog number: A12379 ) (see Recipes)
- Dark phalloidin [Phalloidin from Amanita phalloides (≥90%)] (Sigma-Aldrich, catalog number: P2141 ) (see Recipes)
- Adenosine 5’-triphosphate disodium salt hydrate (ATP) (Sigma-Aldrich, catalog number: A6419 ) (see Recipes)
- Extraction buffer A (see Recipes)
- Extraction buffer B (see Recipes)
- Staining solution (see Recipes)
- Contractility buffer (see Recipes)
Equipment
- Confocal microscope equipped with 100x oil immersion objective
- 37 °C on-stage incubation chamber
Software
- Image J software [National Institutes of Health (NIH)] (http://imagej.nih.gov/ij/)
Procedure
- Substrate patterning and cell seeding
- Micro-contact print circular islands of extracellular matrix protein fibronectin onto 35-mm ibidi’s hydrophobic uncoated μ-dishes to confine cells spreading to an isotropic shape. Circle area of 1,800 μm2 was chosen to ensure cells would spread over the entire island and consistently assemble actin transverse arcs. For a detailed protocol on micro-contact printing see Tee et al. (2015).
- Culture human foreskin fibroblasts (HFF) in growth medium in an incubator at 37 °C with 5% CO2 in air atmosphere.
- To dissociate cells, begin by aspirating culture medium, rinse cells once with 1x PBS and then add trypsin solution to culture flask and incubate for 5 min at 37 °C.
- Resuspend trypsinized cells in growth medium.
- Transfer cells into a centrifuge tube and centrifuge at 800 x g for 3 min.
- Remove supernatant, resuspend cell pellet in growth medium and then centrifuge at 800 x g for 3 min.
- Remove supernatant, resuspend cell pellet in growth medium and then perform cell count.
- Seed 5 x 104 cells in 1 ml growth medium onto the 35-mm printed dish from step A1.
- Incubate dish for 10 min at 37 °C for cells attachment onto fibronectin circular islands.
- Aspirate culture medium and rinse once with 1x PBS to remove unattached cells.
- Add 1 ml of growth medium and return dish to incubator at 37 °C with 5% CO2.
- Incubate cells for 5-7 h prior to cell permabilization.
- Micro-contact print circular islands of extracellular matrix protein fibronectin onto 35-mm ibidi’s hydrophobic uncoated μ-dishes to confine cells spreading to an isotropic shape. Circle area of 1,800 μm2 was chosen to ensure cells would spread over the entire island and consistently assemble actin transverse arcs. For a detailed protocol on micro-contact printing see Tee et al. (2015).
- Cell permeabilization
- Aspirate the culture medium from dish.
- Permeabilize cells by gently adding 1 ml of extraction buffer A.
- Incubate in extraction buffer A for 10 min at room temperature on bench top.
- Gently aspirate extraction buffer A.
- Gently wash permeabilized cells with 2 ml of extraction buffer B.
- Incubate in extraction buffer B for 1-3 min on bench top.
- Gently aspirate extraction buffer B.
- Repeat steps B5-7 three times.
- Aspirate the culture medium from dish.
- Actin cytoskeleton contractility assay
- Stain actin cytoskeleton in cells in 1 ml of staining solution for 10-30 min at room temperature on bench top in the dark.
- [Optional step] Image the AlexaFluor-488-phalloidin labeled actin cytoskeleton in staining solution under 100x objective for 10-30 min at 37 °C with a 2 min time interval and a 5-10 μm Z-section (step-size, 0.35 μm). Actin cytoskeleton should remain inactive prior to induction of contractility with ATP.
- Aspirate staining solution.
- Add 1 ml of contractility buffer.
- Image the AlexaFluor-488-phalloidin labeled actin cytoskeleton in cells under 100x objective for 30-60 min at 37 °C with a 2 min time interval and a 5-10 μm Z-section (step-size 0.35 μm).
- Stain actin cytoskeleton in cells in 1 ml of staining solution for 10-30 min at room temperature on bench top in the dark.
- Kymograph analysis using Image J
- Download freeware Image J from http://imagej.nih.gov/ij/.
- Open time-series Z-stack image (XYZT) in Image J. Perform a Z-projection (click Image → Stacks → Z project), and choose Max Intensity to add up the brightest pixels from each frame. Use the time-series maximum intensity projection image for step D3.
- Draw a line along the region of interest - actin transverse arcs between a pair of radial fibers, using the ‘Straight Line’ tool in Image J. In the example Figure 1, a line is drawn from the cell edge towards the cell center.
- Use the Image J function “Reslice” (click Image → Stacks → Reslice [/]) to generate a kymograph of the time-series stack for the white line using the settings-output spacing 3 pixels, slice count 1, rotate 90 degrees and avoid interpolation. Briefly, each time point gives an intensity line profile, in y-axis, averaged over a 3-pixel width along the drawn line. These line profiles are stacked side by side along the x-axis for all time points, so we get a single image of a distance over time plot in the y- and x-axis respectively (see Figure 1C).
- In the example Figure 1, velocity of the centripetally moving actin transverse arcs can be measured from the slope of the green intensity line (μm min-1) (e.g., in the condition with addition of ATP only). The slope of the line in the kymograph is proportional to velocity. In addition, if the line in the kymograph is parallel to the x-axis (e.g., in the conditions without ATP, with addition of AMP-PNP and addition of ATP together with blebbistatin), this means there is no movement over time.
- Download freeware Image J from http://imagej.nih.gov/ij/.
Representative data
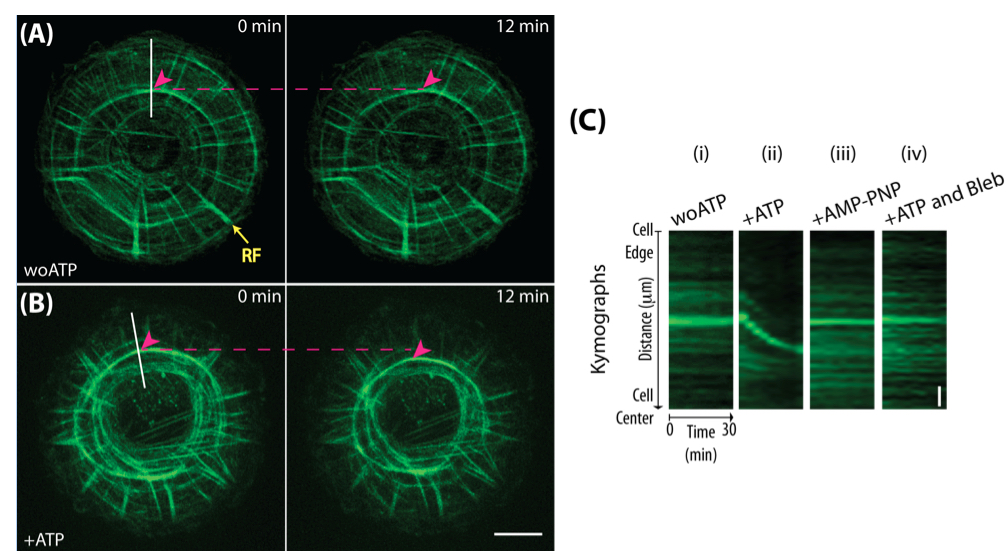
Figure 1. Semi-in vitro detergent-permeabilized cell system. Human foreskin fibroblasts were spread on micro-contact printed circular fibronectin island for 5 h. Actin cytoskeleton (labeled by AlexaFluor-488-phalloidin) organized into a radially symmetrical system with actin radial fibers (RFs) oriented perpendicular to the cell edge and actin transverse arcs (magenta arrowheads) arranged perpendicular to the radial fibers. (A). Actin cytoskeleton remained inactive in contractility buffer without ATP. No movements of the transverse arcs were observed. Yellow arrow points to a radial fiber (RF). (B). Centripetal movement of transverse arcs along the radial fibers was seen in 2 mM ATP containing contractility buffer. Magenta arrowheads indicate the positions of transverse arcs. Magenta dotted lines indicate the initial positions of transverse arcs. Kymograph analysis in C is performed along the white lines indicated. Scale bar, 10 μm. (C). Kymograph analysis of transverse arcs in various experimental conditions: (i) without ATP, (ii) addition of 2 mM ATP, (iii) addition of 2 mM AMP-PNP and (iv) addition of 2 mM ATP together with 100 μM of blebbistatin. Centripetal movement of transverse arcs in permeabilized cells is ATP- and myosin-dependent since movement was only seen following the addition of ATP but not in the conditions with AMP-PNP, a non-hydrolyzable ATP analog, nor in the presence of blebbistatin, a myosin II inhibitor. Vertical scale bar, 2 μm.
Notes
- ATP-induced cell contractility was first reported in water-glycerol extracted cell model by Hoffmann-Berling (1954), see Thery and Piel (2009).
- For an example of actin cytoskeleton contractility assay performed in cells on non-micropatterned glass substrate, see Tint et al. (1991).
- Do not freeze-thaw aliquots of ATP stock solution.
- 2-Mercaptoethanol, phalloidin and protease inhibitors cocktail are to be added fresh each time.
- Addition of PEG and phalloidin to extraction buffer A serve to stabilize the cytoskeleton during extraction.
- Incubation in staining solution and fluorescently-labeled actin cytoskeleton should be kept away from light to minimize bleaching.
- Other fluorescently-conjugated phalloidin can also be used to label the actin cytoskeleton.
- If fluorescent labeling of the actin cytoskeleton is insufficient or excessive, adjust the duration of incubation or dilution of phalloidin in staining solution accordingly.
- If bleaching during image acquisition is significant, add AlexaFluor-488-phalloidin (1:250-500) into the contractility buffer. If the image gets progressively brighter during imaging, reduce the amount of AlexaFluor-488-phalloidin used in the contractility buffer.
- To minimize bleaching during image acquisition, consider increasing camera gain and sensitivity, reducing exposure time and laser intensity and lengthening the time interval between each frame.
- In the event where there is insufficient contrast between the fluorescently-labeled actin cytoskeleton and the background fluorescence from AlexaFluor-488-phalloidin present in the solution during imaging, (i) do not introduce additional fluorescently-tagged phalloidin in contractility buffer and (ii) remove staining solution and wash twice or more times with extraction buffer B prior to imaging control condition in the absence of ATP.
- If drug treatment is needed, drug can be added together with the staining solution prior to induction of ATP-mediated contractility and maintained in the contractility buffer.
- Cell permeabilization can be attempted in cells expressing fluorescently-tagged proteins. In an initial trial, fluorescently-tagged actin markers such as LifeAct-GFP (Riedl et al., 2008) and tdTomato-F-tractin (Johnson and Schell, 2009) are undetectable or weakly seen following cell permeabilization, while fluorescently-tagged protein such as GFP-myosin regulatory light chain and GFP-α-actinin are visible after cell permeabilization. Fluorescence signal retention of other fluorescently-tagged proteins remains to be evaluated.
Recipes
- AlexaFluor-488-phalloidin
Reconstitute in 1.5 ml methanol as per manufacturer instruction
Stored at -20 °C - Dark phalloidin
Reconstitute to 500 μM in ice-cold DMF
Aliquot and stored at -20 °C - ATP
Reconstitute to 500 mM in ice-cold sterile Milli-Q water
Aliquot and stored at -80 °C (see also ‘Notes’ for tips on handling) - Extraction buffer A
50 mM imidazole (pH 6.8)
50 mM KCl
0.5 mM MgCl2
0.1 mM EDTA
1 mM EGTA
1 mM 2-Mercaptoethanol (see also ‘Notes’ for tips on handling)
0.1% Triton-X100
4% PEG MW35000
250 nM dark phalloidin (see also ‘Notes’ for tips on handling)
2 ul ml-1 protease inhibitors cocktail (see also ‘Notes’ for tips on handling) - Extraction buffer B
50 mM imidazole (pH 6.8)
50 mM KCl
0.5 mM MgCl2
0.1 mM EDTA
1 mM EGTA
1 mM 2-Mercaptoethanol
250 nM dark phalloidin
2 μl ml-1 protease inhibitors cocktail - Staining solution
Extraction buffer B, supplement with AlexaFluor-488-phalloidin (1:250 dilution) - Contractility buffer
Extraction buffer B, supplement with 2 mM ATP
Acknowledgments
This protocol was adapted from the previously reported in Tint et al. (1991). This work was supported by the National Research Foundation Singapore, Ministry of Education of Singapore, Grant R-714-006-006-271, and administrated by the National University of Singapore.
References
- Hoffmann-Berling, H. (1954). Adenosintriphosphat als Betriebsstoff von Zellbewegungen. Biochim Biophys Acta 14 (2): 182-194.
- Johnson, H. W. and Schell, M. J. (2009). Neuronal IP3 3-kinase is an F-actin-bundling protein: role in dendritic targeting and regulation of spine morphology. Mol Biol Cell 20(24): 5166-5180.
- Riedl, J., Crevenna, A. H., Kessenbrock, K., Yu, J. H., Neukirchen, D., Bista, M., Bradke, F., Jenne, D., Holak, T. A., Werb, Z., Sixt, M. and Wedlich-Soldner, R. (2008). Lifeact: a versatile marker to visualize F-actin. Nat Methods 5(7): 605-607.
- Tee, Y. H., Shemesh, T., Thiagarajan, V., Hariadi, R. F., Anderson, K. L., Page, C., Volkmann, N., Hanein, D., Sivaramakrishnan, S., Kozlov, M. M. and Bershadsky, A. D. (2015). Cellular chirality arising from the self-organization of the actin cytoskeleton. Nat Cell Biol 17(4): 445-457.
- Tint, I. S., Hollenbeck, P. J., Verkhovsky, A. B., Surgucheva, I. G. and Bershadsky, A. D. (1991). Evidence that intermediate filament reorganization is induced by ATP-dependent contraction of the actomyosin cortex in permeabilized fibroblasts. J Cell Sci 98 (Pt 3): 375-384.
- Thery, M. and Piel, M. (2009). Adhesive micropatterns for cells: a microcontact printing protocol. Cold Spring Harb Protoc 2009(7): pdb prot5255.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Tee, Y. H. and Bershadsky, A. D. (2016). Actin Retrograde Flow in Permeabilized Cells: Myosin-II Driven Centripetal Movement of Transverse Arcs. Bio-protocol 6(5): e1743. DOI: 10.21769/BioProtoc.1743.
Category
Molecular Biology > Protein > Protein-protein interaction
Cell Biology > Cell imaging > Live-cell imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











