- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Post-crystallization Improvement of RNA Crystals by Synergistic Ion Exchange and Dehydration
Published: Vol 5, Iss 17, Sep 5, 2015 DOI: 10.21769/BioProtoc.1578 Views: 9678
Reviewed by: Arsalan DaudiSmita NairAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Preparation of Yeast tRNA Sample for NMR Spectroscopy
Marjorie Catala [...] Pierre Barraud
Jun 20, 2020 5096 Views

Ribosome Purification from an α-proteobacterium and rRNA Analysis by Northern Blot
Liang Yin and Caroline S. Harwood
Dec 5, 2020 3323 Views

In vitro Selection and in vivo Testing of Riboswitch-inspired Aptamers
Michael G. Mohsen and Ronald R. Breaker
Jul 5, 2023 2309 Views
Abstract
Compared to the recent dramatic growth in the numbers of genome-wide and functional studies of complex non-coding RNAs, mechanistic and structural analyses have lagged behind. A major technical bottleneck in the structural determination of large RNAs and their complexes is preparation of diffracting crystals. Empirically, a vast majority of such RNA crystals fail to diffract X-rays to usable resolution (~4 Å) due to their inherent disorder and non-specific packing within the crystals. Here, we present a protocol that combines post-crystallization cation replacement and dehydration that dramatically improved the diffraction quality of crystals of a large gene-regulatory mRNA-tRNA complex. This procedure not only extended the resolution limit of X-ray data from 8.5 to 3.2 Å, but also significantly improved the quality of the data, enabling de novo phasing and structure determination. Because it exploits the general importance of counterions and solvation in RNA structure, this procedure may prove broadly useful in the crystallographic analyses of other large non-coding RNAs.
Keywords: RNA crystallographyMaterials and Reagents
- Oligonucleotides for PCR amplification (IDT DNA Technologies)
- Taq DNA polymerase (5,000 U/ml) (New England Biolabs, catalog number: M0273L )
- T7 RNA polymerase (50,000 U/ml) (New England Biolabs, catalog number: M0251L )
- Diethylpyrocarbonate (DEPC) (catalog number: D5758 )-treated water
Note: DEPC is a toxic alkylating agent and should be handled with appropriate personal protective equipment in a chemical fume hood (Rupert et al., 2004). - KCl (Thermo Fisher Scientific, catalog number: P333-500 )
- MgCl2.6H2O (Sigma-Aldrich, catalog number: M9272-1KG )
- CaCl2.2H2O (Sigma-Aldrich, catalog number: 223506 )
- SrCl2.6H2O (J.T. Baker, catalog number: 4036-01 )
- BaCl2.2H2O (Sigma-Aldrich, catalog number: 217565 )
- UltraPure Low-melting-point agarose (Life Technologies, InvitrogenTM, catalog number: 15517-014 )
- Neutralized tris (2-carboxyethyl) phosphine (TCEP) (Life Technologies, catalog number: 20490 )
- Spermine tetrahydrochloride (Sigma-Aldrich, catalogue number: S1141 )
- Polyethylene Glycol 3350 monodisperse (PEG3350) (Hampton Research, catalog number: HR2-591 )
- Tris base (Thermo Fisher Scientific, catalog number: BP152-10 )
- Boric acid (Thermo Fisher Scientific, catalog number: A73-1 )
- EDTA (Thermo Fisher Scientific, catalog number: BP118-500 )
- Urea (Thermo Fisher Scientific, catalog number: U17-12 )
- RNA binding buffer (see Recipes)
- 20 mM spermine solution (see Recipes)
- Crystallization solution (see Recipes)
- Crystal treatment solutions (see Recipes)
Equipment
- EasyXtal 15-Well Tool (QIAGEN, catalog number: 132007 )
- 9-well glass depression plate (Hampton Research, catalog number: HR3-134 )
- MicroSieves and MicroSaws (MiTeGen, catalog number: T2-L25-A1 )
- 90° angled MicroLoops or MicroMounts (MiTeGen, catalog number: M5-L18SP-A2LD or M2-L18SP-A2 )
- BioRad C1000 Touch PCR thermocycler (Bio-Rad Laboratories, catalog number: 1851197 )
- 37 °C Heat Block (Eppendorf Thermomixer, catalog number: 022670107 )
- Urea polyacrylamide gel electrophoresis system (CBS Scientific custom)
- Handheld UV lamp for UV shadowing (Spectroline, model: EF 140C )
- Whatman Elutrap RNA Electroelution System (GE Healthcare Dharmacon, catalog number: 10447705 )
- Amicon Ultra centrifugal concentrators (10 kD MWCO, 0.5 ml) (Millipore, catalog number: UFC5010BK )
- Leica Stereo microscope (Leica, model: M80 )
Procedure
- Design and synthesis of T-box RNA and tRNA for crystallization
- Initial biochemical and biophysical characterization of T-box RNA-tRNA complexes (Grundy and Henkin, 1993) was essential for design and engineering of crystallization constructs. 20 glycine-specific glyQ/glyQS T-box sequences were selected from a multiple sequence alignment, with preference given to thermophilic, extremophilic, and pathogenic organisms. The choice of glycine-specific T-box system is due to the fact that it is currently the only T-box system that exhibits tRNA-mediated genetic switching with defined components. To aid crystallization, tRNAGly is circularly permuted so that the new 5’ end starts at position 5 in the amino acid acceptor arm, which is capped by a stable, contact-friendly GAAA tetraloop (Zhang and Ferre-D'Amare, 2014a; Zhang and Ferre-D'Amare, 2013; Zhang and Ferre-D'Amare, 2014b).
- The engineered T-box and tRNA are transcribed in vitro using T7 RNA Polymerase and purified by denaturing Urea-PAGE (Milligan and Uhlenbeck, 1989). We typically perform in vitro transcriptions in 2-5 ml volumes and typical yield of purified RNA ranges from 0.2-2 mg per ml of transcription.
- The gel bands containing the RNAs of desired lengths are identified by UV shadowing and are excised using clean razor blades.
- The RNAs are electroeluted from their excised gel pieces using the Whatman Elutrap RNA Electroelution System, in a volume of 500-1,000 μl.
- Wash and concentrate the eluted RNA using an Amicon Ultra spin concentrator (10 kD molecular weight cut off, 0.5 ml). Each centrifugation run is at ~14,000 rcf and lasts 8-12 min, or until the retentate volume is less than 100 μl. Wash the RNA once with 1 M KCl and three times with DEPC-treated water, concentrate it and store it at 4 °C or -20 °C before use (Klein and Ferre-D'Amare, 2006).
- Initial biochemical and biophysical characterization of T-box RNA-tRNA complexes (Grundy and Henkin, 1993) was essential for design and engineering of crystallization constructs. 20 glycine-specific glyQ/glyQS T-box sequences were selected from a multiple sequence alignment, with preference given to thermophilic, extremophilic, and pathogenic organisms. The choice of glycine-specific T-box system is due to the fact that it is currently the only T-box system that exhibits tRNA-mediated genetic switching with defined components. To aid crystallization, tRNAGly is circularly permuted so that the new 5’ end starts at position 5 in the amino acid acceptor arm, which is capped by a stable, contact-friendly GAAA tetraloop (Zhang and Ferre-D'Amare, 2014a; Zhang and Ferre-D'Amare, 2013; Zhang and Ferre-D'Amare, 2014b).
- Crystallization of the T-box stem I-tRNA-YbxF ternary complex
- Dilute ~40 μl concentrated tRNAGAAA (~1 mM; 24 g/L) to ~20 μM using 1,960μl DEPC-treated water to reduce intermolecular interaction and dimerization.
- “Snap-cool” tRNAGAAA by incubating at 90 °C for 3 min followed by rapid cooling to 4 °C using a PCR thermocyler, using the fastest ramping rate available (5 °C/sec as used). Alternatively, after heating to 90 °C for 3 min, tRNA is rapidly chilled by putting it on ice immediately.
- Concentrate refolded tRNA to ~12 g/L (500 μM) using centrifugal concentrators (10 kD MWCO; 0.5 ml). Each centrifugation run is at ~14,000 rcf and lasts 8-12 min, or until the retentate volume is less than 100 μl.
- Mix in a total volume of ~20 μl to produce 200 μM final concentrations of each T-box Stem I RNA and snap-cooled tRNA in 1x RNA binding buffer (see Recipes), incubate first at 50 °C for 10 min and then at 37 °C for 30 min on a PCR thermocycler or heat block. Use heated lid when possible to reduce condensation and alterations of sample volume.
- Add one equivalent (~200 µM final) selenomethionyl Bacillus subtilis (B. subtilis) YbxF (Baird et al., 2012) to the mRNA-tRNA binary complex to form a 1:1:1 ternary complex in 10-40 µl total volume. Incubate at room temperature for 2 min.
- Add spermine to a final concentration of 2 mM. The mixture may become temporarily cloudy but should quickly clarify. Mix gently by pipetting up and down gently.
- Melt a stock of 2% low-melting-point agarose solution on a 90 °C heat block and allow it to slowly cool to 37 °C on a heat block to prevent it from solidifying. The agarose solution can be stored at 4 °C for at least a month and reused multiple times.
- Mix 1:1 the sample solution of the T-box ternary complex and crystallization solution (see Recipes), keep at 37 °C.
- Add 1/10 volumes of 2% low-melting-point agarose solution and gently mix by pipetting up and down. The presence of agarose fibers in crystal solvent channels lends mechanical support to the crystals (Biertumpfel et al., 2002; Lorber et al., 2009). In this case, the presence of 0.2% low-melting-point agarose effectively protects the co-crystals from severe cracking due to the sudden changes in osmolarity in the dehydration step. A range of agarose concentrations from 0.05% to 0.5% have been tested, yielding a variety of consistencies ranging from fluid liquid (<0.1%) to viscous liquid (~0.1%), to jelly-like solid (~0.2%) to robustly solid (>0.2%). In our experience, the jelly-like solid consistency (~0.2%) permits easy handling of the drop, facilitates excision of crystals from the agarose network, and provides sufficient mechanical support for the crystals.
- Fill the reservoirs of the EasyXtal 15-Well Tool with 300-500 μl of crystallization solution.
- Transfer the crystallization mixture (1-2 μl in total volume) onto the cover slides of the EasyXtal 15-Well Tool.
- Tightly screw the cover slides onto the EasyXtal 15-Well Tool plate to commence crystallization experiments by hanging drop vapor diffusion.
- Dilute ~40 μl concentrated tRNAGAAA (~1 mM; 24 g/L) to ~20 μM using 1,960μl DEPC-treated water to reduce intermolecular interaction and dimerization.
- Post-crystallization treatments
- Square-plate-shaped crystals of the T-box-tRNA-YbxF ternary complex start appearing in a few days. Diffraction quality crystals tend to grow more slowly, reaching final dimensions of 320 x 300 x 60 μm3 over the course of 3-4 weeks. These crystals belong to space group C2221, with unit cell dimensions of a=108.7 Å, b=108.8 Å, c=291.8 Å. As do many other plate-shaped macromolecular crystals with one relatively longer unit cell edge, the c axis of these crystals is collinear with their shortest physical dimension, i.e., the edge, or “rim” that describes the thickness of the rectangular or rhombic plates. Thus, oscillation diffraction images that result from incident X-rays that traverse through the broad faces of the plates suffer from significant overlap of adjacent reflections. Such overlap can be circumvented by the use of 90° bent crystal loops (custom bending of MiTiGen loops), which restrict the incident X-rays to only enter and exit the crystals through their rims but not their faces of the plates.
- Depending on the final concentration of low-melting point agarose in the crystallization drop and temperature, the entire drop may exhibit a number of consistencies ranging from fluid, to viscous, to jelly-like, to robustly solid. Appropriate crystal-manipulating tools are selected accordingly to transfer crystals into 100-200 μl Crystal Treatment Solutions in glass depression plates (Equipment). Use conventional nylon loops to transfer single crystals from non-viscous liquid drops, and use tools such as MicroSieves (MiTeGen) to transfer whole, solidified drops. Harvesting from viscous, soft jelly-like drops can be inefficient and technically challenging, and is thus best avoided when possible. A gradient of concentrations of the primary precipitant (20-50% PEG3350 in 5% increments) is scouted to achieve a range of final solvent contents and the effect on diffraction quality is measured. Different concentrations of a panel of divalent cations especially the alkaline earth metals (Mg2+, Ca2+, Sr2+, Ba2+) are screened, both for supporting crystal growth and for post-crystallization treatment. In the case of the T-box complex crystals, crystal grown in the presence of Mg2+ and subsequently treated post-crystallization in Sr2+ stands out as the optimal procedure (Table 1), producing circular, well-separated Bragg spots. Such high quality data were essential for de novo phasing of the complex structure using single-wavelength anomalous dispersion (SAD), taking advantage of the two selenomethinines present in the YbxF protein.
- Seal each well of the depression plate using a square glass cover slide and Vaseline. Incubate the crystals in crystal treatment solution (see Recipes) at room temperature for 16 h. For the crystals of the T-box ternary complex, shorter treatments (i.e. less than 4 h) generally do not produce the full effect of the prolonged treatment. Generally, if the crystals exhibit excessive physical damage from a treatment, one may mitigate the shock to the crystals by serially transferring the crystals through several intermediate crystal treatment solutions that have incremental changes in composition. In the case of the T-box co-crystals, the single-step “shock” treatment produced better results as compared to several step-wise treatments, aided by the mechanical support from the agarose network.
- Carefully dissect the crystals out from their surrounding agarose network using MicroSaws (MiTeGen) and remove as much as agarose as possible without physically damaging the crystals (Figure 1). As the orientation of the crystals in the crystal loop is critical for reducing overlap during data collection, it is essential to trim nearly all agarose away from the crystal faces so that the plate-like crystals would be mounted parallel to the plane of the 90° bent loops. We recommend the use of microtools made of soft-touch materials such as plastic or polymer, rather than metal tools. The involuntary vibrations from the hand holding a metal spatula tend to cause more severe physical damage to the crystals. “Dual-wielding” two plastic MiTeGen MicroSaws under the stereomicroscope appears to afford confident manipulations while minimizing localized physical impact.
- Using a 90° bent loop such as the angled MicroLoops or MicroMounts (MiTeGen), pick up single, trimmed crystals and immediately plunge into liquid nitrogen for vitrification. As the Crystal Treatment Solution already contains at least 40% (w/v) polyethylene glycol (PEG) 3350, no additional cryoprotective agent is necessary.
- Square-plate-shaped crystals of the T-box-tRNA-YbxF ternary complex start appearing in a few days. Diffraction quality crystals tend to grow more slowly, reaching final dimensions of 320 x 300 x 60 μm3 over the course of 3-4 weeks. These crystals belong to space group C2221, with unit cell dimensions of a=108.7 Å, b=108.8 Å, c=291.8 Å. As do many other plate-shaped macromolecular crystals with one relatively longer unit cell edge, the c axis of these crystals is collinear with their shortest physical dimension, i.e., the edge, or “rim” that describes the thickness of the rectangular or rhombic plates. Thus, oscillation diffraction images that result from incident X-rays that traverse through the broad faces of the plates suffer from significant overlap of adjacent reflections. Such overlap can be circumvented by the use of 90° bent crystal loops (custom bending of MiTiGen loops), which restrict the incident X-rays to only enter and exit the crystals through their rims but not their faces of the plates.
- Understanding treatment-induced improvement of crystal quality
- Structure determination of as-grown, untreated crystals, and a number of crystals subjected to various combinations of post-crystallization treatments permitted the tracking of macromolecular movements in these crystals in response to the treatments administered (Table 1) (Zhang and Ferre-D'Amare, 2013; Zhang and Ferre-D'Amare, 2014b).
- Structural alignment of untreated and optimally treated crystals reveal that the ternary complexes of T-box-tRNA-YbxF undergo rotations and translations as quasi-rigid bodies to closer proximity to each other in the crystal, producing superior packing contacts including several intimate base-stacking interactions between symmetry-related complexes as well as a Class-I A-minor interaction between the engineered RNA tetraloop that caps the tRNA acceptor stem and the minor groove of the proximal region of T-box Stem I (Zhang and Ferre-D'Amare, 2014b).
- The unique effect of Sr2+ in post-crystallization treatments of T-box co-crystals may be in part explained by its specific association with the 3' cis-diols of neighboring symmetry-related T-box RNAs, its frequent bidentate innersphere interactions with the Hoogsteen faces of purines, its binding to bulges and junctions where phosphates cluster, or bridging across the narrow major groove on A-form RNA helices. The ability of Sr2+ to bind RNA 3' termini and its flexible coordination geometry are properties that may allow it to improve crystalline packing of RNA (Hofer et al., 2006).
- Structure determination of as-grown, untreated crystals, and a number of crystals subjected to various combinations of post-crystallization treatments permitted the tracking of macromolecular movements in these crystals in response to the treatments administered (Table 1) (Zhang and Ferre-D'Amare, 2013; Zhang and Ferre-D'Amare, 2014b).
Representative data
Table 1. Select properties of crystals treated with varying degrees of ion replacement and dehydration
| PDB code | Li2SO4 (mM) | MgCl2 (mM) | SrCl2 (mM) | PEG 3350 (% w/v) | Resolution (Å) | Space Group | Unit cell dimensions (Å) | VM (Å3/Da) | VS (%) |
| 4TZP | 300 | 20 | 0 | 20 | 8.5 | C2221 | 108.7, 108.8, 291.8* | 3.26 | 74.6 |
| 4TZV | 0 | 20 | 0 | 20 | 5.0 | P43212 | 75.7, 75.7, 270.2* | 2.93 | 71.7 |
| 4TZW | 0 | 0 | 50 | 20 | 4.7 | P43212 | 75.3, 75.3, 268.9* | 2.89 | 71.3 |
| 4TZZ | 0 | 100 | 0 | 48 | 3.6 | P21 | 70.6, 260.7, 70.7 † | 2.46 | 66.3 |
| 4LCK | 0 | 0 | 40 | 40 | 3.2 | C2221 | 100.8, 109.7, 268.1* | 2.81 | 70.4 |
*α = ß = Γ = 90°
†α = Γ = 90°, ß = 92.8°
VM Matthews coefficient
Vs Calculated solvent content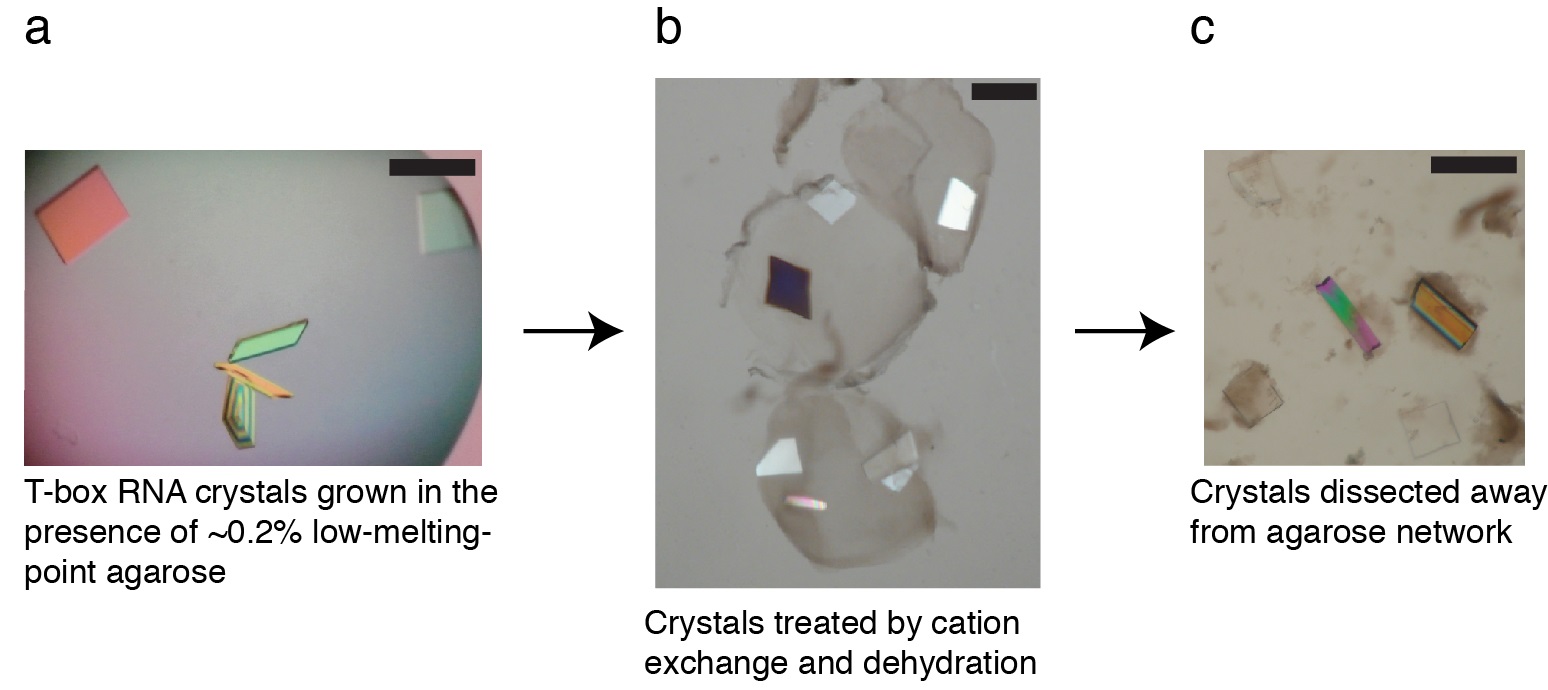
Figure 1. Appearances of T-box RNA crystals after initial growth in the presence of agarose (a), during treatment (b), and after dissection (c). Solid bar indicates 200 μm.
Recipes
- RNA binding buffer (1x)
50 mM HEPES-KOH (pH 7.0)
100 mM KCl
20 mM MgCl2
5 mM tris (2-carboxyethyl) phosphine (TCEP) - 20 mM spermine solution
In DEPC-treated water
Filtered through 0.2 mm filter - Crystallization solution
50 mM Bis-Tris (HCl) (pH 6.5)
0.3 M Li2SO4
20 mM MgCl2
20% (w/v) polyethylene glycol (PEG) 3350 - Crystal treatment solutions
50 mM Bis-Tris (HCl) (pH 6.5)
100 mM KCl
20–50 mM SrCl2 or 20-100 mM MgCl2
40-45% PEG3350
5 mM TCEP
Acknowledgments
We thank the staff at beamlines 5.0.1 and 5.0.2 of the ALS and ID-24-C and ID-24-E of APS, in particular, K. Perry and K. R. Rajashankar of the Northeastern Collaborative Access Team (NE-CAT) of the APS for support in data collection and processing, and N. Baird, C. Jones, M. Lau, A. Roll-Mecak, and K. Warner for discussions. This work is partly based on research conducted at the ALS on the Berkeley Center for Structural Biology beamlines and at the APS on the NE-CAT beamlines (supported by National Institute of General Medical Sciences grant P41GM103403). Use of ALS and APS was supported by the U.S. Department of Energy. J. Zhang is a recipient of the NHLBI Career Transition Award (K22). This work was supported in part by the intramural program of the NHLBI, NIH.
References
- Baird, N. J., Zhang, J., Hamma, T. and Ferre-D'Amare, A. R. (2012). YbxF and YlxQ are bacterial homologs of L7Ae and bind K-turns but not K-loops. RNA 18(4): 759-770.
- Biertumpfel, C., Basquin, J., Suck, D. and Sauter, C. (2002). Crystallization of biological macromolecules using agarose gel. Acta Crystallogr D Biol Crystallogr 58(Pt 10 Pt 1): 1657-1659.
- Grundy, F. J. and Henkin, T. M. (1993). tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell 74(3): 475-482.
- Hofer, T. S., Randolf, B. R. and Rode, B. M. (2006). Sr(II) in water: A labile hydrate with a highly mobile structure. J Phys Chem B 110(41): 20409-20417.
- Klein, D. J. and Ferre-D'Amare, A. R. (2006). Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science 313(5794): 1752-1756.
- Lorber, B., Sauter, C., Theobald-Dietrich, A., Moreno, A., Schellenberger, P., Robert, M. C., Capelle, B., Sanglier, S., Potier, N. and Giege, R. (2009). Crystal growth of proteins, nucleic acids, and viruses in gels. Prog Biophys Mol Biol 101(1-3): 13-25.
- Milligan, J. F. and Uhlenbeck, O. C. (1989). Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol 180: 51-62.
- Rupert, P. B. and Ferre-D'Amare, A. R. (2004). Crystallization of the hairpin ribozyme: illustrative protocols. Methods Mol Biol 252: 303-311.
- Zhang, J. and Ferre-D'Amare, A. R. (2013). Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA. Nature 500(7462): 363-366.
- Zhang, J. and Ferre-D'Amare, A. R. (2014a). New molecular engineering approaches for crystallographic studies of large RNAs. Curr Opin Struct Biol 26: 9-15.
- Zhang, J. and Ferre-D'Amare, A. R. (2014b). Dramatic improvement of crystals of large RNAs by cation replacement and dehydration. Structure 22(9): 1363-1371.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Zhang, J. and Ferré-D’Amaré, A. R. (2015). Post-crystallization Improvement of RNA Crystals by Synergistic Ion Exchange and Dehydration. Bio-protocol 5(17): e1578. DOI: 10.21769/BioProtoc.1578.
Category
Biochemistry > RNA > RNA structure
Molecular Biology > RNA > RNA structure
Molecular Biology > RNA > RNA synthesis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











