- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Virus-induced Gene Silencing (VIGS) in Phalaenopsis Orchids
Published: Vol 4, Iss 24, Dec 20, 2014 DOI: 10.21769/BioProtoc.1359 Views: 13909
Reviewed by: Tie LiuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

RNA Stability Measurements Using RT-qPCR in Arabidopsis Seedlings
Tianran Jia and Brandon H. Le
Jul 20, 2020 6996 Views

Laser-Assisted Microdissection and High-Throughput RNA Sequencing of the Arabidopsis Gynoecium Medial and Lateral Domains
Valentín Luna-García and Stefan de Folter
Sep 5, 2024 2066 Views

Effective Gene Silencing in Plants by Synthetic Trans-Acting siRNAs Derived From Minimal Precursors
Adriana E. Cisneros [...] Alberto Carbonell
Oct 20, 2025 1806 Views
Abstract
This is a protocol to produce stable silencing efficacy and efficiency for VIGS using CymMV as a silencing vector for floral functional genomics in Phalaenopsis orchids. This protocol is established based on a method created by Lu et al. (2007), and then modified by Hsieh et al. (2013a; 2013b), Lu et al. (2012) successfully engineered a cloning vector (pCymMV-Gateway) in that the target gene fragment is simple to insert and can be manipulated with high efficiency. The silencing vector is inoculated into plants by Agro-inoculation by using Agrobacterium tumefaciens (A. tumefaciens) strain EHA105. Agro-infiltration of leaves for use in VIGS study of orchid flowers is a time saver and produces less damage to flower buds.
Materials and Reagents
- Virus-free Phalaenopsis plants
- TOP10 Escherichia coli (E. coli) competent cells
- Agrobacterium tumefaciens strain EHA105
- Gel/PCR DNA fragments extraction kit (Geneaid Biotech, catalog number: DF100 )
- Gateway® BP-ClonaseTM II enzyme mix (Life Technologies, InvitrogenTM, catalog number: 11789-020 )
- Selective plate (LB plate with 50 µg/ml Kanamycin)
- High-speed plasmid mini kit (Geneaid Biotech, catalog number: PD100 )
- Acetosyringone (Sigma-Aldrich, catalog number: D134406 )
- Murashige and Skoog salt (Sigma-Aldrich, catalog number: M5524 )
- Tryptone (US Biological, catalog number: 12855 )
- Yeast extract (Affymetrix, catalog number: 23547 )
- NaCl (Sigma-Aldrich, catalog number: S1446 )
- Tris-HCl (US Biological, catalog number: 22676 )
- CaCl2 (Inter-County Mechanical Corp., catalog number: 03-11250 )
- Glycerol (J.T.Baker®, catalog number: 2136-01 )
- Luria Broth medium (LB medium) (see Recipes)
- Proteinase K solution (see Recipes)
- Murishige and Skoog medium (MS medium) salt (see Recipes)
Equipment
- GeneAmp PCR system 9700 (Life Technologies, Applied Biosystems®)
- Laminar flow cabinet
- BioChrom Libra S50 UV/Vis Spectrophotometer (Biochrom)
- Incubator shaker
- Electroporation machine (Kaneka Corporation, Eurogentec)
- Electroporation cuvettes (EquiBio, catalog number: ECU-102 )
- Centrifuges for MiniSpin Eppendorf and 50 ml conical centrifuge tubes
- 1-ml syringe with a needle
- Water bath
Procedure
- Construction of pCymMV-Gateway plasmid DNA
- DNA fragments for silenced target gene for insertion into pCymMV-Gateway vector (Figure 1A) is obtained by PCR amplification with oligonucleotide primers containing 29-nt attB recombination site in both ends (see Note 1). Purify attB-PCR products by using a gel extraction kit.
- Insert target gene fragment into pCymMV-Gateway vector by using BP recombination reaction (Figure 1B).
- In a 1.5 ml microcentrifuge tube at room temperature mix attB-PCR product (150 ng), pCymMV-Gateway vector (150 ng), and 4 µl of 5x BP ClonaseTM reaction buffer. Then add 1x TE buffer (pH 8.0) until a total volume of 16 µl is obtained.
- Remove the BP ClonaseTM enzyme mix from -80 °C storage and put in ice for 2 min to thaw and vortex briefly twice (2 sec each time).
- Add 4 µl of BP ClonaseTM enzyme mix to each reaction (step A2a, above). Mix well by vortexing twice and then microcentrifuge briefly. Store BP ClonaseTM enzyme mix at -80 °C immediately after usage.
- Incubate reactions at 25 °C for 60 min.
- Add 2 µl of the Proteinase K solution to each sample to terminate the reaction.
- Vortex briefly and incubate samples at 37 °C for 10 min.
- In a 1.5 ml microcentrifuge tube at room temperature mix attB-PCR product (150 ng), pCymMV-Gateway vector (150 ng), and 4 µl of 5x BP ClonaseTM reaction buffer. Then add 1x TE buffer (pH 8.0) until a total volume of 16 µl is obtained.
- Transform competent E. coli with the pCymMV-Gateway vector for amplification (in the Laminar flow cabinet).
- Incubate TOP10 E. coli competent cells on ice for 10 min (see Note 2).
- Add 5 µl of each BP reaction (step A2) into 50-100 µl of TOP10 E. coli competent cells.
- Incubate on ice for 15 min.
- Heatshock cells by incubating at 42 °C water bath for 45 sec.
- Incubate on ice for 2 min.
- Add 200 µl of LB medium and incubate at 37 °C for 60 min while shaking at 200 rpm.
- Spread each transformation (305 µl) onto LB-agar medium plates containing 50 µg/ml of Kanamycin.
- Incubate at 37 °C for 13 h.
- Incubate TOP10 E. coli competent cells on ice for 10 min (see Note 2).
- Randomly select two to four colonies for each construct, and then subculture.
- Recover and purify the plasmids.
- Confirm each construct by PCR with the gene-specific forward primer and the general reverse primer (CymMV 5351, 5′-CTTCTGTACCATACACATAG-3′) on the gateway vector.
- DNA fragments for silenced target gene for insertion into pCymMV-Gateway vector (Figure 1A) is obtained by PCR amplification with oligonucleotide primers containing 29-nt attB recombination site in both ends (see Note 1). Purify attB-PCR products by using a gel extraction kit.
- Transformation by electroporation after amplification of the recombined pCymMV-Gateway vector (steps B1-3 in the Laminar flow cabinet)
- Thaw the Agrobacterium tumefaciens (strain EHA105) competent cells at room temperature and immediately place on ice. Transfer 100 μl of the competent cells to a chilled electroporation cuvette on ice. Add 1-2 μl of recombined pCymMV-Gateway vector (150 ng of plasmid DNA can be used depending on the competence of the cells), mix gently and keep on ice.
- Perform the electroporation.
- Insert the cuvettes in the electroporation chamber and initiate the electroporation. The voltage and pulse time parameters are pre-set.
- Immediately add 200 μl of LB medium in the cuvettes and transfer the solution to a sterile Eppendorf tube.
- Incubate the mixture at 28 °C for 30 min. while shaking at 200 rpm.
- Insert the cuvettes in the electroporation chamber and initiate the electroporation. The voltage and pulse time parameters are pre-set.
- Spread all the mixture on selected LB-agar medium plate containing 100 μM of Acetosyringone and 50 µg/ml of Kanamycin and incubate at 28 °C for 16 h.
- Confirm each colony by PCR with the gene-specific forward primer and the general reverse primer (CymMV 5351, 5′-CTTCTGTACCATACACATAG-3′) on the gateway vector for the presence of the modified constructs.
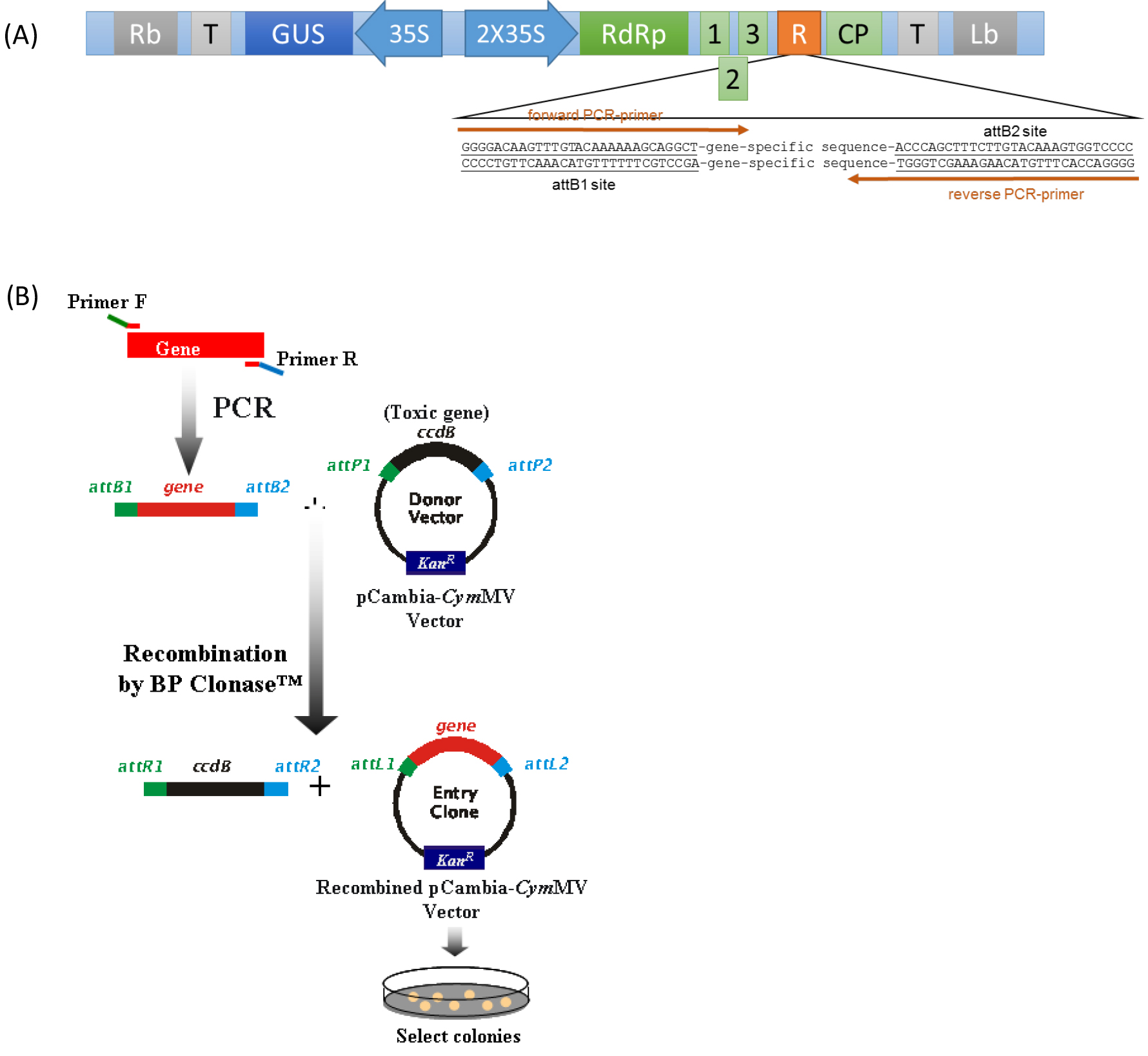
Figure 1. (A) Schematic representation of pCymMV-Gateway vector. Green rectangles represent open reading frames encoded by Cymbidium mosaic virus (CymMV) genomic RNA. RNA-dependent RNA polymerase (RdRp); triple gene block 1, 2, and 3; coat protein (CP) and attB sites are indicated. (B) Amplification of target gene and construction of pCambia-CymMV vector.
- Thaw the Agrobacterium tumefaciens (strain EHA105) competent cells at room temperature and immediately place on ice. Transfer 100 μl of the competent cells to a chilled electroporation cuvette on ice. Add 1-2 μl of recombined pCymMV-Gateway vector (150 ng of plasmid DNA can be used depending on the competence of the cells), mix gently and keep on ice.
- Agro-infiltration of Phalaenopsis plants
- Prepare electroporation competent Agrobacterium cells:
- Four days prior to infection, transfer a single colony of Agrobacterium tumefaciens containing recombinant plasmids into 5 ml of LB medium containing 100 μM of Acetosiringone and 50 µg/ml of kanamycin and culture at 28 °C for 16 h while shaking at 200 rpm.
- Subculture the bacterial cell in 50 ml of LB medium containing 100 μM of Acetosiringone and 50 µg/ml of kanamycin and incubate at 28 °C for 13-16 h while shaking at 200 rpm until OD600 reaches 0.8-1.0 (about 3-5 h).
- To harvest, transfer the culture to 50 ml centrifuge bottles and centrifuge for 10 min at 4 °C 3,000 x g. From this step the cells should be kept cold throughout the preparation.
- After centrifugation, remove as much of the supernatant as possible. Resuspend cell pellets by adding 300 µl of MS medium containing 100 µM of Acetosyringone. It is allowed to stand at room temperature for 0.5 hours without shaking before agro-infiltration.
- Four days prior to infection, transfer a single colony of Agrobacterium tumefaciens containing recombinant plasmids into 5 ml of LB medium containing 100 μM of Acetosiringone and 50 µg/ml of kanamycin and culture at 28 °C for 16 h while shaking at 200 rpm.
- Infiltrate 100 µl of Agrobacterium tumefaciens containing recombined pCymMV Gateway vector in virus-free Phalaenopsis plants by use of a 1-ml syringe with a needle (see Note 3).
- Perform infiltration using either one of the two methods (Figures 2 and 3):
- For leaf injection, inject suspensions into the abaxial side of blade or laminar regions but excluding form tip, base, midrib, and margin parts of the leaf (usually the third youngest leaf for Phalaenopsis) right above the inflorescence emerge (Figure 2, left panel and Figure 3A).
- For inflorescence injection, inject suspensions into the floral stalk (Figure 2, right panel and Figure 3B) of the raceme with eight internodes and one visible floral bud (extruding out of its bract). The raceme stalk usually emerges from the stem between the third and the fourth leaves.
After injection, the injection areas become dark green under the epidermal surface than the un-injected ones (Figure 3C-D).
- For leaf injection, inject suspensions into the abaxial side of blade or laminar regions but excluding form tip, base, midrib, and margin parts of the leaf (usually the third youngest leaf for Phalaenopsis) right above the inflorescence emerge (Figure 2, left panel and Figure 3A).
- Plants after injected should be grown back in stable and regular growth condition in an insect-proof and thermal-controlled (20-25 °C) greenhouse.
- Agro-infiltrated inflorescences bloom with the first blooming flower appearing on 31-35 days post Agro-infiltration.
- Confirm Agrobacterium infiltration, virus movement and silencing of transgene expression by PCR (see Note 4).
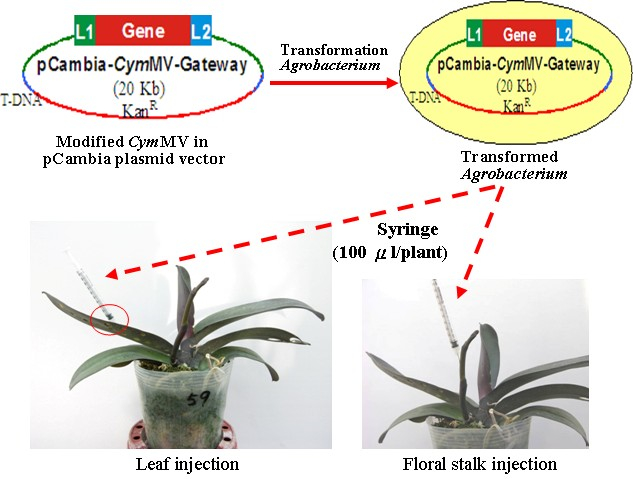
Figure 2. Agrobacterium infiltration in virus-free Phalaenopsis plants. A. The Agrobacteriums with the recombined pCambia-CymMV vectors are injected into the leaf (left panel) or the floral stalk (right panel) with Agro liquid culture using a 1-milliliter syringe.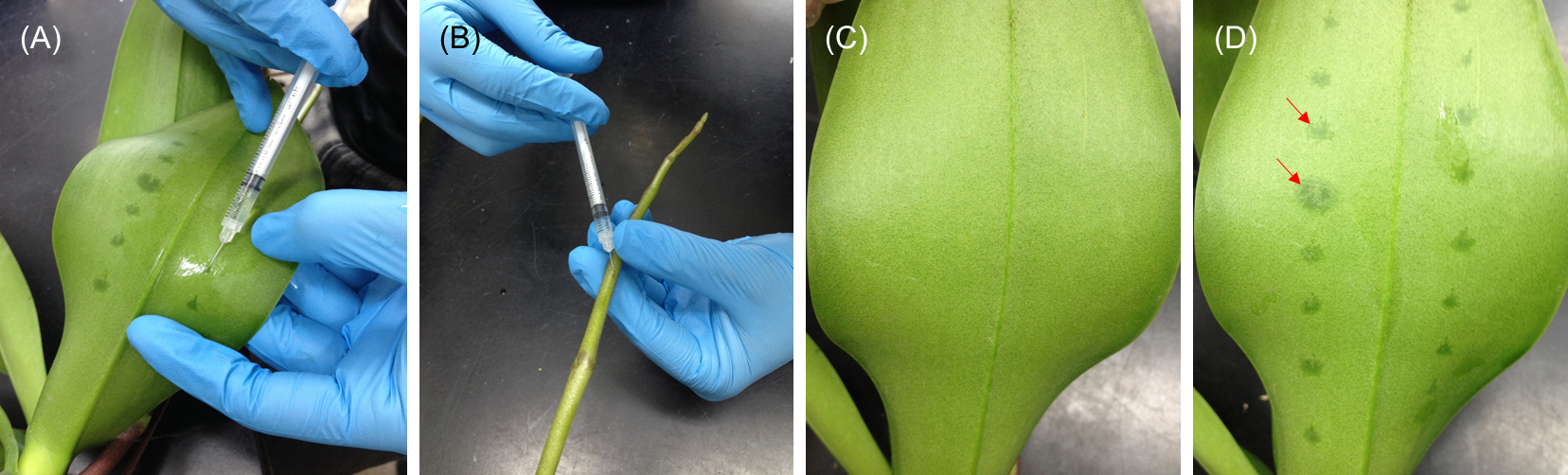
Figure 3. Demonstration of performing infiltration in (A) abaxial side of leaf and (B) floral stalk. (C and D) The leaves before and after injection. The injection areas can be distinguishably observed (red arrows).
- Prepare electroporation competent Agrobacterium cells:
Notes
- To generate PCR products suitable for use as substrates in a Gateway® BP recombination reaction with a pCymMV-Gateway vector, the attB sites are needed to be incorporated into your PCR products:
- The forward PCR-primer must contain the attB1 site (5′ GGGGACAAGTTTGTACAAAAAAGCAGGCT-3′) at the 3′ end followed by at least 18-25 bp of template- or gene-specific sequences.
- The reverse PCR-primer must contain the attB2 site (5′-GGGGACCACTTTGTACAAGAAAGCTGGGT-3′) at the 3′ end followed by at least 18-25 bp of template- or gene-specific sequences.
- Remember that the gene-specific nucleotides need to be in frame with the attB sequence and that stop codons should be removed.
- The position and length of amplicon of the insert fragments used for silencing are different depending on individual genes and need to be optimized for each target genes. e.g. Both large (1,498 bp) and small (81 bp) sizes of insert fragments of PeUFGT3 showed significant silencing effects (Hsieh et al., 2013a). To avoid non-specific targeting, it is better to select gene-specific region for primer design.
- For insert fragment amplification, standard PCR reaction condition is used, but it is better to use DNA polymerase with proof reading ability. A higher agarose percentage enhances resolution of smaller bands to distinguish PCR products from the primers by agarose gel electrophoresis. Elute DNA in d.d.H2O or TE buffer after purify PCR products by using a gel extraction kit.
- The forward PCR-primer must contain the attB1 site (5′ GGGGACAAGTTTGTACAAAAAAGCAGGCT-3′) at the 3′ end followed by at least 18-25 bp of template- or gene-specific sequences.
- The transformation efficiency of TOP10 E. coli competent cells should be equal to or exceed 1.0 x 109 colonies/ml.
- The optimal amount of Agrobacteirum mixture for infiltration depends on the growth stage, organ and variety of Phalaenopsis plants. We recommend infiltrating 100 µl of Agrobacteirum mixture in each mature Phalaenopsis plants to ensure that their development and growth are not affected.
- cDNA synthesis was performed with total RNAs extracted from leaf or floral tissues above injected regions. RT-PCR was performed with the gene-specific forward primer and the generalreverseprimer (CymMV 5351, 5′-CTTCTGTACCATACACATAG-3′) on the gateway vector.
Recipes
- Luria Broth medium (LB medium)
10 g/L tryptone
5 g/L yeast extract
10 g/L NaCl (pH 7.0) - Proteinase K solution
2 μg/μl Proteinase K in 10 mM Tris-HCl (pH 7.5)
20 mM CaCl2
50% glycerol - Murishige and Skoog medium (MS medium) salt
Major salts (macronutrients):
1,650 mg/L ammonium nitrate (NH4NO3)
440 mg/L calcium chloride (CaCl2.2H2O)
370 mg/L magnesium sulphate (MgSO4.7H2O)
170 mg/L potassium phosphate (KH2PO4)
1,900 mg/L potassium nitrate (KNO3) Minor salts (micronutrients)
6.2 mg/L boric acid (H3BO3)
0.025 mg/L cobalt chloride (CoCl2.6H2O)
0.025 mg/L cupric sulphate (CuSO4.5H2O)
27.8 mg/L ferrous sulphate (FeSO4.7H2O)
22.3 mg/L manganese sulphate (MnSO4.4H2O)
0.83 mg/L potassium iodide (KI)
0.25 mg/L codium molybdate (Na2MoO4.2H2O)
8.6 mg/L zinc sulphate (ZnSO4.7H2O)
37.2 mg/L Na2EDTA.2H2O
pH 5.8
Acknowledgments
The authors would like to thank Dr. Lu, H. C. (Biotechnology Center in Southern Taiwan, Academia Sinica) for his assistance in viral vector construction.
References
- Hsieh, M. H., Lu, H. C., Pan, Z. J., Yeh, H. H., Wang, S. S., Chen, W. H. and Chen, H. H. (2013a). Optimizing virus-induced gene silencing efficiency with Cymbidium mosaic virus in Phalaenopsis flower. Plant Sci 201-202: 25-41.
- Hsieh, M. H., Pan, Z. J., Lai, P. H., Lu, H. C., Yeh, H. H., Hsu, C. C., Wu, W. L., Chung, M. C., Wang, S. S., Chen, W. H. and Chen, H. H. (2013b). Virus-induced gene silencing unravels multiple transcription factors involved in floral growth and development in Phalaenopsis orchids. J Exp Bot 64(12): 3869-3884.
- Lu, H. C., Chen, H. H., Tsai, W. C., Chen, W. H., Su, H. J., Chang, D. C. and Yeh, H. H. (2007). Strategies for functional validation of genes involved in reproductive stages of orchids. Plant Physiol 143(2): 558-569.
- Lu, H. C., Hsieh, M. H., Chen, C. E., Chen, H. H., Wang, H. I. and Yeh, H. H. (2012). A high-throughput virus-induced gene-silencing vector for screening transcription factors in virus-induced plant defense response in orchid. Mol Plant Microbe Interact 25(6): 738-746.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Hsieh, M., Pan, Z., Yeh, H. and Chen, H. (2014). Virus-induced Gene Silencing (VIGS) in Phalaenopsis Orchids. Bio-protocol 4(24): e1359. DOI: 10.21769/BioProtoc.1359.
Category
Plant Science > Plant molecular biology > RNA > RNA interference
Plant Science > Plant molecular biology > RNA > Transcription
Molecular Biology > RNA > RNA interference
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








