- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In situ Digestion of Wheat Cell-wall Polysaccharides
Published: Vol 4, Iss 23, Dec 5, 2014 DOI: 10.21769/BioProtoc.1306 Views: 12605
Reviewed by: Arsalan DaudiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Enzymatic Starch Quantification in Developing Flower Primordia of Sweet Cherry
Nestor Santolaria [...] Afif Hedhly
Apr 5, 2025 1885 Views

New Approach to Detect and Isolate Rhamnogalacturonan-II in Arabidopsis thaliana Seed Mucilage
Dayan Sanhueza and Susana Saez-Aguayo
Sep 5, 2025 1240 Views

Detailed Method for the Purification of Rhamnogalacturonan-I (RG-I) in Arabidopsis thaliana
Liang Zhang [...] Breeanna R. Urbanowicz
Feb 5, 2026 99 Views
Abstract
Cell walls of the wheat endosperm are mostly composed of arabinoxylans (AX) and mixed (1→3), (1→4)-β-glucans (BG) (Saulnier et al., 2012). Here, we present an optimized protocol to degrade enzymatically these cell-wall polysaccharides into oligosaccharides, directly from wheat grain cross sections. The main difficulty is to provide a sufficient amount of humidity for the enzyme to be active, while the amount of liquid at the surface of the tissue should stay low to prevent any delocalization of the released products. With this protocol, enzymatic degradation was shown to be efficient and delocalization of released oligosaccharides was estimated below 50 µm (Veličković et al., 2014).
Although it can be employed for other purposes, this in situ enzymatic digestion was primarily developed to obtain molecular images of the cross-sections of wheat endosperm by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry (Veličković et al., 2014). The cell wall polysaccharides are heterogeneous in structure, exhibit high masses and are entangled into complex networks. Thus, they are not amenable to direct analysis by mass spectrometry and they need to be degraded into smaller compounds as a first step. In this protocol, additional steps corresponding to the deposition of the MALDI matrix are also described.
Materials and Reagents
- Milli-Q quality water (dH2O)
- Ethanol (Sigma-Aldrich, catalog number: 02854 )
- Acetonitrile (Sigma-Aldrich, catalog number: 271004 )
- K2SO4 (Merck KGaA, catalog number: 5153 )
- NaCl (Merck KGaA, catalog number: 106404.1 )
- CaCl2.2H2O (CARLO ERBA Reagents, catalog number: 433381 )
- NaN3 (Merck KGaA, catalog number: 822335 )
- NaOH (Sigma-Aldrich, catalog number: 221465 )
- H3PO4 (Sigma-Aldrich, catalog number: P6560 )
Enzymes
- α-Amylase from porcine pancreas (Sigma-Aldrich, catalog number: A3176 )
- Xylanase M1 from Trichoderma viride (Megazyme International, catalog number: E-XYTRI )
- Lichenase [endo-1,3(4)-β-D-glucanase] from Bacillus sp. (Megazyme International, catalog number: E-LICHN )
MALDI matrix preparation
- 2,5-dihydroxybenzoic acid (DHB) (Sigma-Aldrich, catalog number: 85707 )
- N,N-dimethylaniline (DMA) (analytical reagent grade) (Thermo Fisher Scientific, catalog number: 121-69-7 )
Buffers and media
- 70% ethanol (see Recipes)
- 0.1 M NaOH (see Recipes)
- 25 mM CaCl2 (see Recipes)
- Buffer for α-Amylase (see Recipes)
- 1 mg/ml α-Amylase (see Recipes)
- 460 U/ml Xylanase M1 (see Recipes)
- 4.6 U/ml Xylanase M1 (see Recipes)
- 200 U/ml Lichenase (see Recipes)
- 2 U/ml Lichenase (see Recipes)
- Saturated K2SO4 at 40 °C (see Recipes)
- 50% acetonitrile (see Recipes)
- MALDI matrix preparation (see Recipes)
Equipment
- Vibratome (MICROM, model: HM 650 V )
- In-house designed spraying robot
Note: Robot was built by adapting an electro-spray probe dismounted from a LCQ Advantage mass spectrometer (Thermo Fisher Scientific) to an X, Y, Z robotic arm (FISNAR, F4300N) (see Figure 1) (see Note 2).
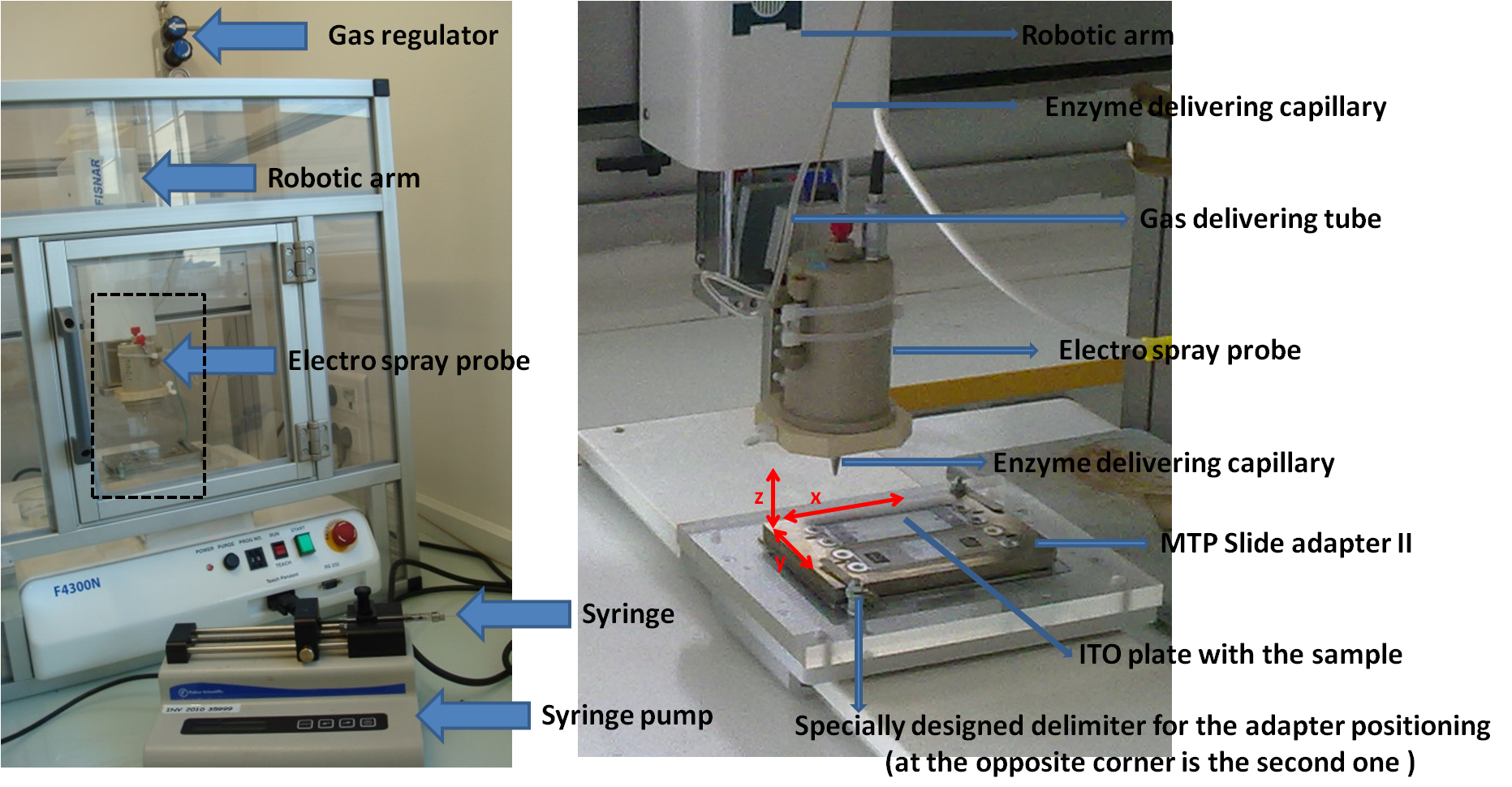
Figure 1. In-house designed spraying robot for enzyme application. The right hand panel represents the enlarged view of the dashed rectangle zone depicted on the left picture.
- Syringe pump infusion 0.2 µl/h to 500 ml/h (Thermo Fischer Scientific, catalog number: 12486350 )
- 500 µl glass syringe (Hamilton, model: Gastight 1750 )
- Thermo-shaker (Eppendorf, Thermomixer compact)
- Incubator (THERMOSI, model: SR 300 )
- Home-designed chamber for incubation
Note: This consists of a rubber gasket sealed glass container (the jar with lid, KORKEN, IKEA of Sweden. Diameter 11 cm; high 10.5 cm, volume 0.5 L) in which is placed a 50 ml glass beaker and a set of weights (e.g. pieces of stones) (see Figure 2). Set of weights are required to keep the glass beaker from floating.
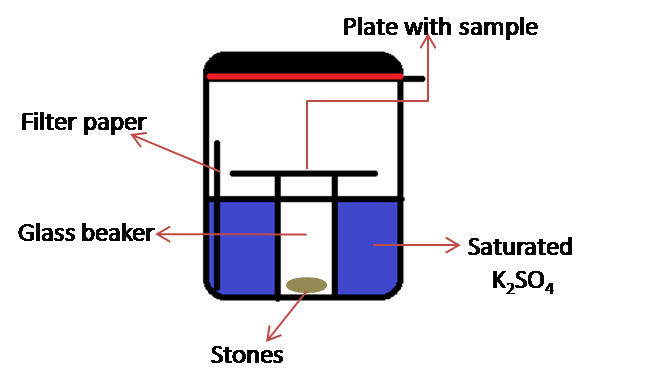
Figure 2. Schematic representation of home-designed chamber for incubation
- pH meter (pHenomenal®, model: pH 1000 L , equipped with a pHenomenal® 221 electrode)
Other equipment
- Glue (Loctite Super Glue Ultra Gel)
- Adhesive carbon tape (8 mm x 20 m) (Agar Scientific, catalog number: AGG3939 )
- Indium tin oxide (ITO) glass slides (Bruker, catalog number: 237001 ) (see Note 3)
- Art paint brush (approximate diameter of the brush: 3-4 mm)
- Razor blade (Gillette, Bleue extra)
- Petri dish (90 mm x 14.2 mm) (Thermo Fischer Scientific, catalog number: 5184E )
- Whatman filter paper (90 mm diameter)
- 1.5 ml Eppendorf tube (Eppendorf Safe-Lock quality)
- 2 ml plastic Pasteur pipette (Thermo Fischer Scientific, catalog number: 13984 )
- Kimwipes paper (Kimberly-Clark)
Additional, required for MALDI imaging experiments
- ImagePrep nebulizing robot (Bruker Daltonics) (see Figure 3)
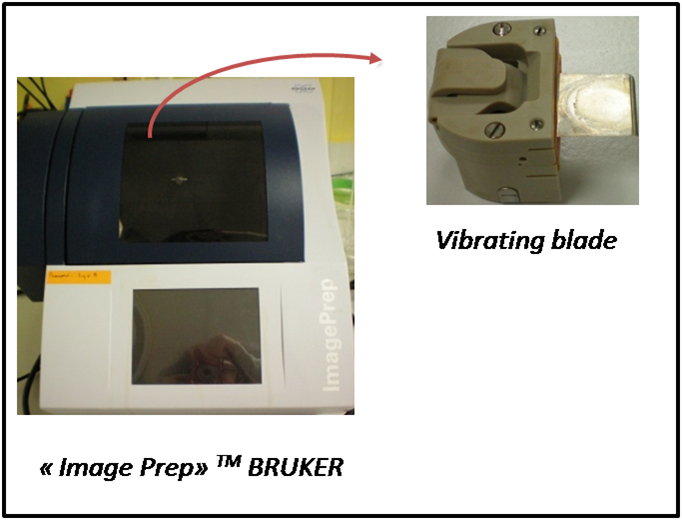
Figure 3. ImagePrep nebulizing robot
- MTP Slide adapter II (Bruker Daltonics, catalog number: 235380 ) (see Note 4)
Procedure
- Rehydration of wheat grains (Philippe et al., 2006).
- The germ part (see Figure 4) is removed with a razor blade, perpendicular to the crease zone (longitudinal axis) of the grain, and is discarded.
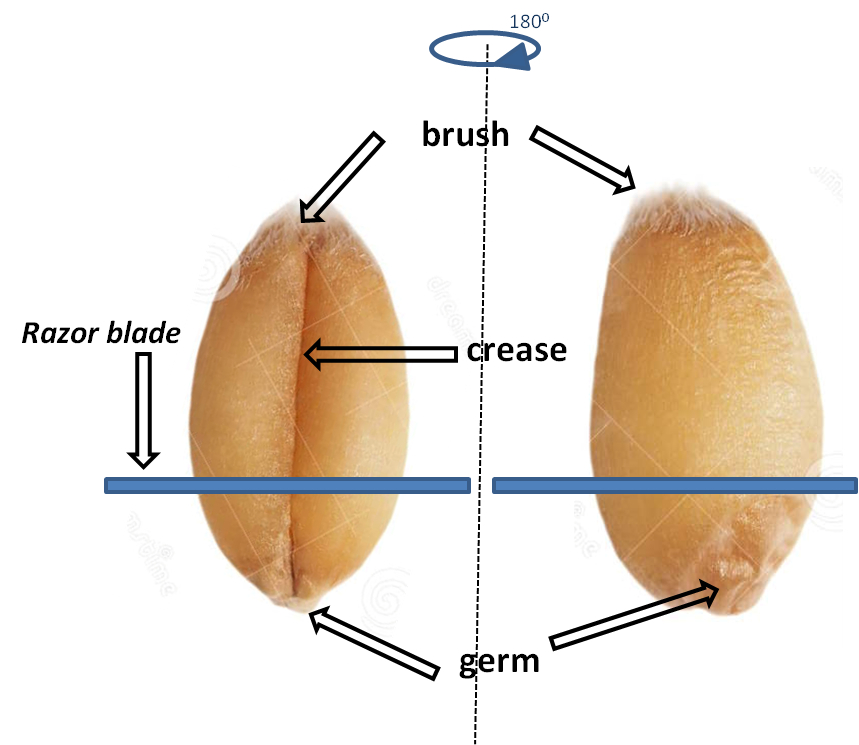
Figure 4. Wheat grain and position of razor blade during wheat germ removal. Both sides of grain are presented to easily localize the germ, brush and crease regions.
- The grain is then placed in a Petri dish on top of Whatman filter paper moistened with dH2O, and left at +4 °C for 24 h.
Note: Whole rehydration procedure is skipped for young grains, which are naturally hydrated.
- The germ part (see Figure 4) is removed with a razor blade, perpendicular to the crease zone (longitudinal axis) of the grain, and is discarded.
- Preparing 70 µm-thin cross sections of wheat grains.
- This step is done by using a Vibratome instrument according to instruction provided in the Instruction manual (Thermo Fisher Scientific, 2009). The grain is fixed with some glue onto the magnetic plate of the Vibratome. Orientation of the grain should be such that the longitudinal axis of the grain is placed perpendicular to the magnetic plate. Settings of the Vibratome instrument are as follows: frequency: 60; amplitude: 1.2; speed: 12.
- A series of 400 µm-thick slices are cut and discarded, until the surface of the grain becomes flat and parallel to the razor blade of the Vibratome instrument.
- Thin sections (approximately 70 µm) are then cut consecutively and placed immediately into Eppendorf tubes filled with 70% ethanol. They are stored at 4 °C until further processing.
- This step is done by using a Vibratome instrument according to instruction provided in the Instruction manual (Thermo Fisher Scientific, 2009). The grain is fixed with some glue onto the magnetic plate of the Vibratome. Orientation of the grain should be such that the longitudinal axis of the grain is placed perpendicular to the magnetic plate. Settings of the Vibratome instrument are as follows: frequency: 60; amplitude: 1.2; speed: 12.
- Removing starch.
- The wheat cross section is grabbed using a paint brush and transferred into a clean Eppendorf tube, filled with 0.5 ml of 1 mg/ml α-Amylase solution. This is allowed to incubate at 40 °C for 24 h on a Thermo-shaker.
- Rinse thoroughly by transferring the wheat cross section into a clean Eppendorf tube filled with 1 ml of dH2O. Repeat this step twice. Wash the brush in water after each sample transfer.
Note: This step is intended to avoid any hydrolysis of starch by endogenous enzyme activity, which would produce some glucans of similar masses as those expected from the hydrolysis of cell walls BG (additionally, when MALDI mass spectrometry is used for further analysis of the tissue, starch generates a strong suppressive effect on the signal).
- The wheat cross section is grabbed using a paint brush and transferred into a clean Eppendorf tube, filled with 0.5 ml of 1 mg/ml α-Amylase solution. This is allowed to incubate at 40 °C for 24 h on a Thermo-shaker.
- Placing the cross-section onto an ITO glass plate (see Note 5).
- Prepare the ITO glass plate that will receive the cross section:
- Place a small piece of adhesive carbon tape at the location where the cross section will be deposited on the ITO glass plate;
- With a Pasteur pipette, put a droplet of water on the tape.
- Place a small piece of adhesive carbon tape at the location where the cross section will be deposited on the ITO glass plate;
- Then, with a paint brush, grab the cross section from the Eppendorf tube. Lean the cross section at the surface of the water droplet, allowing it to detach from the paint brush. Proceed very gently, so to avoid any disruption or any distortion of the cross section (Figure 5).
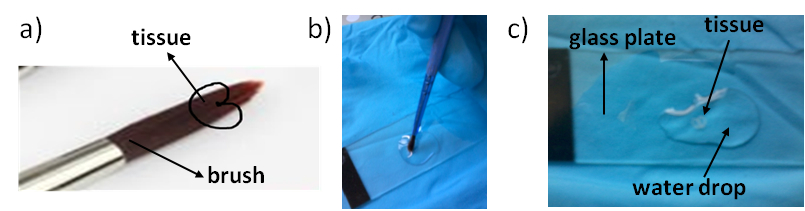
Figure 5. Positioning of grain section during wheat section grabbing a) and deposition onto the ITO plate (b, c).
Gently remove excess water with a Kimwipes tissue. Allow sample to dry at RT (approximately 30 min) (see Figure 6).
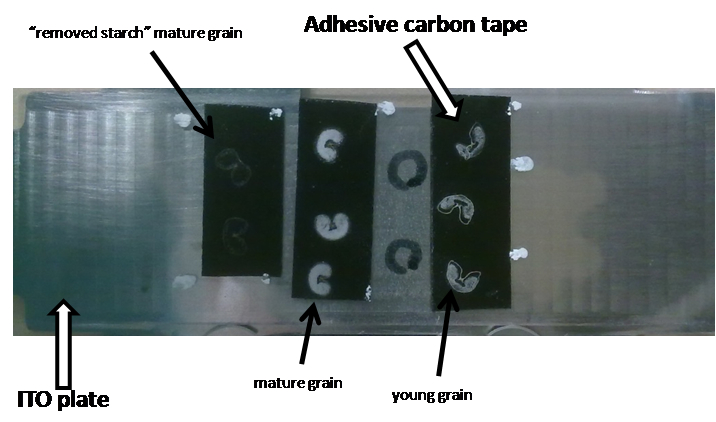
Figure 6. Appearance of the ITO plate with mounted wheat sections on it
- Mount the ITO glass plate onto the MTP Slide Adapter II (Bruker) according to the manufacturer instructions [available in Bruker, (2012)].
- Prepare the ITO glass plate that will receive the cross section:
- Application of the hydrolytic enzyme on the cross section.
- This step uses a homemade robot (see Figure 1 and Note 2). The aim is to deliver a controlled volume of enzyme onto the tissue by using an airbrush device, so that fine droplets of enzyme are applied on the tissue, thereby limiting the diffusion of the oligosaccharides released upon digestion over the tissue.
- Spraying of the enzyme is achieved by connecting the electrospray probe mounted on the robotic arm to a syringe pump delivering the enzymatic solution at a constant flow rate of 600 μl/h.
- Spraying is assisted pneumatically with nitrogen (1.5 x 105 Pa).
- The distance between the needle tip of the Electrospray probe and the ITO plate is 3 cm (Z-axis).
- A X, Y deposition pattern following a “brush rectangle” is used (Figure 7).
- The movement speed of the robot head was set at 5 mm/s.
- Other parameters (X and Y axis start and end coordinates, volume of the enzyme placed into the syringe, number of spraying cycles) are set to ensure that the robot consistently deposits 0.3 μl of enzyme (4.6 U/ml endo-1,4-β-xylanase or 2 U/ml lichenase) per mm2 of sprayed area (corresponding to 0.0014 U xylanase and 0.0006 U lichenase per mm2 tissue).
Enzymes can be applied individually, consecutively or in mixture.
Note: Endo-1,4-β-xylanase is used for AX hydrolysis, while lichenase is used for mixed-linkage BG hydrolysis.
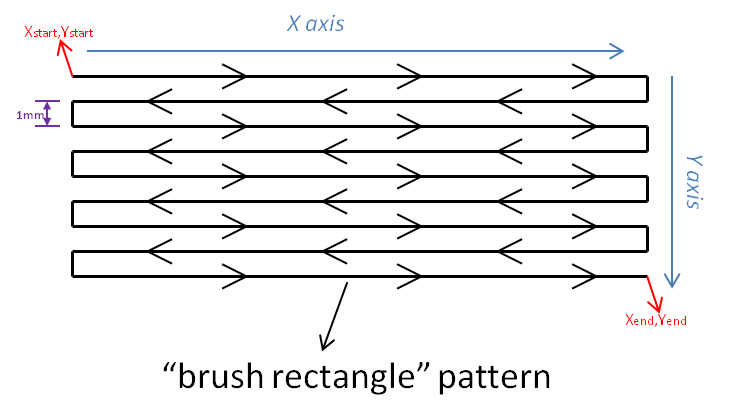
Figure 7. “Brush rectangle” pattern used for enzyme deposition. X and Y “start” and “end” positions are determined from the coordinates of the surface which must be covered by the enzyme.
- Spraying is assisted pneumatically with nitrogen (1.5 x 105 Pa).
- This step uses a homemade robot (see Figure 1 and Note 2). The aim is to deliver a controlled volume of enzyme onto the tissue by using an airbrush device, so that fine droplets of enzyme are applied on the tissue, thereby limiting the diffusion of the oligosaccharides released upon digestion over the tissue.
- Incubation.
- The wheat cross sections covered by the enzyme in the previous step are allowed to incubate at 40 °C for 4 h.
- To prevent liquid from evaporating too fast and the enzyme becomes inactive, a wet atmosphere is maintained by placing the ITO plate into a sealed incubation chamber filled with 150 ml saturated K2SO4 and pre-incubated at 40 °C.
- The ITO plate is placed on top of a 50 ml glass beaker weighted with stones and installed into the glass incubator (Figure 2).
- After incubation, remove the tissues from the incubation chamber and let dry in the open air (approximately 15 min).
- The wheat cross sections covered by the enzyme in the previous step are allowed to incubate at 40 °C for 4 h.
- Additional step for MALDI MS measurement of the hydrolyzed cross sections: Deposition of the DMA-DHB MALDI matrix.
- This step is performed with an automatic vibration vaporization system from Bruker (ImagePrep, see Figure 3) according to the instructions provided by the manufacturer (Bruker Daltonik, 2007).
- The settings are as follows (see Note 6): 1st Phase: 15 cycles: 12% spray power; 20% spray modulation; 2 sec spray time; 15 sec incubation time; 30 sec dry time. 2nd Phase: 40 cycles: 20% spray power; 25% spray modulation; 2 sec spray time; 30 sec incubation time; 60 sec dry time. During whole procedure N2 flow is provided at 2 x 105 Pa.
- This step is performed with an automatic vibration vaporization system from Bruker (ImagePrep, see Figure 3) according to the instructions provided by the manufacturer (Bruker Daltonik, 2007).
Representative data
- Figure 8 gives a representative example of the type of results which can be expected by this method. The localization of released AX oligosaccharides after in-tissue hydrolysis by endo-1,4-β-xylanase is depicted, as measured by MALDI MS imaging.
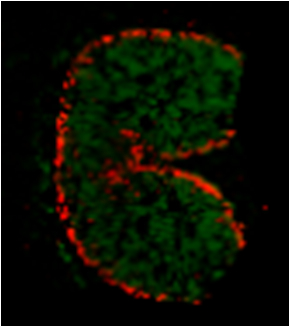
Figure 8. MALDI MS imaging of released AX oligosaccharides at the wheat cross section after endo-1,4-β-xylanase in-situ hydrolysis. Red pixels indicate places where feruloylated AX of DP5 (degree of polymerization 5) is present and green pixels indicate non-feruloylated AX oligosaccharides of DP5 and DP6.
Recipes
- 70% ethanol
Place 700 ml ethanol in a glass bottle and complete to 1,000 ml with dH2O
- 0.1 M NaOH
Weigh 0.4 g NaOH and dissolve in 100 ml of dH2O
- 25 mM CaCl2
Weigh 0.018 g CaCl2.2H2O and dissolve in 5 ml of dH2O
- Buffer for α-Amylase (20 mM Na-phosphate buffer with 2 mM NaCl and 0.25 mM CaCl2, pH 6.9 with 0.02% NaN3 as stabilizer)
Place 450 ml of dH2O in a glass bottle, and add
1.15 g H3PO4
0.058 g NaCl
5 ml of 25mM CaCl2
Mix thoroughly and adjust pH to 6.9 with 0.1 M NaOH
Complete to 500 ml with dH2O
Add 0.1 g NaN3
Stored at +4 °C
- One mg/ml α-Amylase
Dissolve 10 mg of α-Amylase in 10 ml of buffer for α-Amylase
- 460 U/ml Xylanase M1
Place 80 μl of dH2O in a clean Eppendorf tube
Add 20 μl of manufacture stock of Xylanase M1 (2,300 U/ml)
Gently shake before sampling
- 4.6 U/ml xylanase M1
Place 990 μl of dH2O in a clean Eppendorf tube
Add 10 μl of 460U/ml Xylanase M1
- 200 U/ml lichenase
Place 80 μl of dH2O in a clean Eppendorf tube.
Add 20 μl of manufacture stock of lichenase (1,000 U/ml)
Gently shake before sampling
- 2 U/ml lichenase
Place 990 μl of dH2O in a clean Eppendorf tube
Add 10 μl of 200 U/ml lichenase
- Saturated K2SO4 at 40 °C
Weigh 75 g K2SO4 and dissolve in 500 ml of dH2O (warm up to 40 °C with stirring until complete dissolution)
- 50% acetonitrile
Place 250 ml of acetonitrile in a glass bottle
Complete to 500 ml with dH2O
- MALDI matrix preparation
Weigh 500 mg DHB
Dissolve in 5 ml of 50% acetonitrile
Add 100 μl of DMA
Careful: DMA is toxic in contact with skin and suspected of causing cancer. Wear protective gloves, protective clothing, eye protection and face protection. Proceed under a fume hood.
Notes
- This protocol was performed on mature wheat grains (700 D) from several cultivars of the genus Triticum aestivum: Recital, Malacca, Virtuose, Magdalena, Crousty, Thesee, Baltimore, Sisley, Aligre and Tamaro.
The same protocol was also performed on young stages of development (245 D) from seeds of the cultivar Recital.
- The basic idea of the home-made robot is to put an airbrush-type device on a X, Y, Z robotic arm, and be able to control the liquid flow rate through the airbrush. To do so, we have dismounted an Electrospray probe from an old LCQ Advantage mass spectrometer and adjusted this probe to a FISNAR 4300N robot. Flow rate through the probe is controlled by a syringe pump, delivering a typical flow rate of 1-100 µl/min. Nitrogen is used as a co-axial nebulizing gas, at a pressure of 1.5 x 105 Pa.
The FISNAR 4300N robot is controlled by a Teach Pendant device, which enables to program the motion patterns for the enzyme deposition. A program consists in defining the start and end positions (defined by X, Y, Z coordinates), the pattern and the speed of the robotic arm movement. “Brush rectangle” pattern is one of the available patterns and is the one that we used in our experiments. It is depicted in Figure 7. The programing and use of the FISNAR 4300N robot is very well explained in (FISNAR, 2012).
- ITO glass plates: These plates exhibit a conductive side (coated by Indium Tin Oxide). The use of these conductive plates was imposed in our case by the MALDI mass spectrometry measurements that were subsequently made on the hydrolyzed tissues. However, any glass plate with the same dimensions (75 x 25 x 0.9 mm) can be used in principle.
- Adapter for glass slides (MTP Slide adapter II for glass slides): This adapter is provided by Bruker in order to introduce ITO glass plates into an Autoflex III MALDI mass spectrometer. It was used herein as a convenient holder for all the steps performed with the homemade spraying robot. However, any rectangular holder of the same dimensions (12.8 x 8.8 cm) can be used instead.
- Note that the most critical step for maintaining reproducibility of in-tissue digestion is transferring the tissue at the ITO plate (Procedure 4). It must be very carefully performed to avoid any damage of the tissue cell wall network. Several sections (3 to 4) must usually be deposited so to ensure that at least one of these is of good quality (this is also dependent of the wheat cultivar and stage of development of the seed. There is no other trick than experience!). Application of the enzyme is performed automatically by the robot so the reproducibility of this step is high (does not depend on the position of the section on the plate). It is however recommended that a fresh enzyme solution is prepared for this step. Our experience is that these points are the main reason of result variance (which was estimated to be of 14% with our detection method based on MALDI MS).
- The ImagePrep device produces an aerosol by vaporization of the MALDI matrix through a vibration blade, i.e. a thin sheet of stainless steel with pinholes. These holes can get clogged or expand after several uses. The settings (spray power, e.g.) can be adjusted by the operator to ensure a similar coating of the tissue with the MALDI matrix, even though the blade has been used several times. However, we recommend replacing the vibrational blade after ten uses (or whenever it looks damaged) to ensure reproducible application of the MALDI matrix.
Acknowledgments
This work was supported in part through a postdoctoral fellowship (Dušan Veličković) from INRA (Institut National de Recherche Agronomique, France) and AgreenSkills. We are very grateful to Fabienne Guillon and Luc Saulnier (INRA Biopolymers, Interaction, Assemblies, Nantes, France) for helpful discussions about the enzymatic hydrolysis of wheat cell walls. This protocol is a modified version of the protocol previously described in Veličković et al. (2014).
References
- Bruker Daltonic (2007). Image Prep User Manual.
- Bruker Daltonic (2012). Instructions for Use Tools for MALDI Imaging.
- FISNAR (2012). F4000N Series Robot Operating Manual.
- Philippe, S., Robert, P., Barron, C., Saulnier, L. and Guillon, F. (2006). Deposition of cell wall polysaccharides in wheat endosperm during grain development: Fourier transform-infrared microspectroscopy study. J Agric Food Chem 54(6): 2303-2308.
- Saulnier, L., Guillon, F. and Chateigner-Boutin, A. L. (2012). Cell wall deposition and metabolism in wheat grain. J Cereal Sci 56(1): 91-108.
- Thermo Fisher Scientific (2009). Thermo Scientific Microtome with vibrating blade MICROM HM 650V: Instruction manual.
- Veličković, D., Ropartz, D., Guillon, F., Saulnier, L. and Rogniaux, H. (2014). New insights into the structural and spatial variability of cell-wall polysaccharides during wheat grain development, as revealed through MALDI mass spectrometry imaging. J Exp Bot 65(8): 2079-2091.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Veličković, D. and Rogniaux, H. (2014). In situ Digestion of Wheat Cell-wall Polysaccharides. Bio-protocol 4(23): e1306. DOI: 10.21769/BioProtoc.1306.
Category
Plant Science > Plant biochemistry > Carbohydrate
Plant Science > Plant physiology > Tissue analysis
Biochemistry > Carbohydrate > Polysaccharide
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









