- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Binding Affinity Measurement of Antibodies from Crude Hybridoma Samples by SPR
Published: Vol 4, Iss 21, Nov 5, 2014 DOI: 10.21769/BioProtoc.1276 Views: 17336
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Determination of Dissociation Constants for the Interaction of Myosin-5a with its Cargo Protein Using Microscale Thermophoresis (MST)
Rui Zhou [...] Xiang-Dong Li
Feb 5, 2025 1738 Views

Cell-Sonar, an Easy and Low-cost Method to Track a Target Protein by Expression Changes of Specific Protein Markers
Sabrina Brockmöller [...] Simone Rothmiller
Feb 5, 2025 1657 Views

SiMPull-POP: Quantification of Membrane Protein Assembly via Single Molecule Photobleaching
Ryan J. Schuck [...] Rajan Lamichhane
Jan 5, 2026 287 Views
Abstract
Surface Plasmon Resonance (SPR) is widely used to generate kinetic and affinity information on specific interactions between biomolecules. This technique is label-free and monitors the binding event in real-time. It is generally used for characterization of monoclonal antibody - antigen interactions. This protocol describes specifically the use of SPR with a Biacore T100 instrument to measure the affinity of crude hybridoma samples to a protein. For that purpose an anti-IgG antibody was firstly covalently immobilized onto a CM5 chip by amide coupling (Canziani et al., 2004; Schraml and Biehl, 2012). Then the antibodies from hybridoma supernatants were captured non-covalently onto the surface via their Fc region providing an optimal analyte-binding orientation. Finally, the resulting complex was stabilized by crosslinking with EDC/NHS to avoid baseline drift during measurement and regeneration (Pope et al., 2009). Then the interaction with the protein was monitored at several concentrations and its affinity towards the immobilized antibodies was determined with the corresponding KD obtained from classical kinetics analysis. This set-up avoids the avidity effects of the bivalent antibodies, allows the use of non-purified analytes with unknown concentrations and the specific capture of the antibodies in a similar stable covalent-orientated manner.
Materials and Reagents
- Dulbecco’s phosphate buffered saline (D-PBS) (Sigma-Aldrich, catalog number: D8537 ) as running buffer during immobilization and binding analysis
- Amine coupling kit (GE Healthcare, catalog number: BR-1000-50 ) containing:
- 750 mg N-ethyl-N’-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC)
- 115 mg N-hydroxysuccinimide (NHS)
- 10.5 ml Ethanolamine-HCl (pH 8.5)
- 750 mg N-ethyl-N’-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC)
- Mouse antibody capture kit (GE Healthcare, catalog number: BR-1008-38 )
- 10 mM acetate buffer (pH 5) (GE Healthcare, catalog number: BR-1003-51 )
- 10 mM acetate buffer (pH 5.5) (GE Healthcare, catalog number: BR-1003-52 )
- 10 mM Glycine-HCl (pH 1.7) (GE Healthcare, catalog number: BR-1008-38) as regeneration buffer
- Reagents for immobilization (see Recipes)
- Ligands (see Recipes)
- Reagents for binding assay (see Recipes)
- Analyte (see Recipes)
Equipment
- Sensor chip with carboxyl groups available for the amine coupling reaction: Serie S sensor chip CM5 (GE Healthcare, catalog number: BR-1005-30 )
- Instrument for SPR analysis: Biacore T100 instrument and Evaluation Software (GE Healthcare)
- Micropipettes (100-1,000 µl) (Eppendorf, catalog number: 033023B )
- Micropipettes (10-100 µl) (Eppendorf, catalog number: 033021B
- Micropipettes (2-20 µl) (Eppendorf, catalog number: 033019B )
- Plastic vials (0.8 ml) (GE Healthcare, catalog number: BR-1002-12 )
- Rubber caps (type 3) (GE Healthcare, catalog number: BR-1005-02 )
Software
- Biacore T100 Evaluation Software (GE Healthcare)
Procedure
- Immobilization of the antibodies from hybridoma supernatants
- Prepare 167 µl of 30 µg/ml anti-mouse IgG solution in the 10 mM acetate buffer (pH 5).
- The capture reagent anti-mouse IgG then was immobilized in four flow cells (fc) using the amino coupling protocol and following the instructions described in the “Mouse antibody capture kit” using the PBS buffer as running buffer. At the end of the anti-mouse IgG immobilization, the regeneration solution was injected three times to remove any anti-mouse IgG non-covalent bound to the chip, leaving intact the covalent bound anti-mouse IgG.
Note: Flow cells are formed when a microfluidic flow system is brought into contact with a sensor surface on which the ligand is immobilized. Here, the sample is injected separately through independent, unconnected flow cells, although other configurations as serial flow cells or hydrodynamic addressing flow cell systems are possible.
The reagents and parameters used for the different steps (activation of the carboxymethyl groups of the chip surface, ligand immobilization, surface deactivation and regeneration) are compiled in Table 1.
Note: An immobilization level of anti-mouse IgG around 9,000-10,000 Resonance Unit (RU) is usually obtained.
Table 1. Immobilization procedure of the anti-mouse IgG antibodyNotes: *Flow cell 1 serves as a reference cell and is only immobilized with the anti-mouse IgG.Cycle Reagent Flow cell (fc) Contact time (sec) Flow rate (µl/min) 1 1:1 (v/v) mixture of EDC (0.4 M) and NHS (0.1 M) 1*, 2, 3, 4 420 5 µl/min Anti-mouse IgG 1*, 2, 3, 4 420 5 µl/min 1 M Ethanolamine-HCl (pH 8.5) 1*, 2, 3, 4 420 5 µl/min 2** 10 mM Glycine-HCl (pH 1.7) 1*, 2, 3, 4 3 times 60 50 µl/min
**This step is done in a different cycle and it is not shown in the Figure 1 below. - An example of the sensorgram obtained for the anti-mouse IgG immobilization is shown in Figure 1 below.
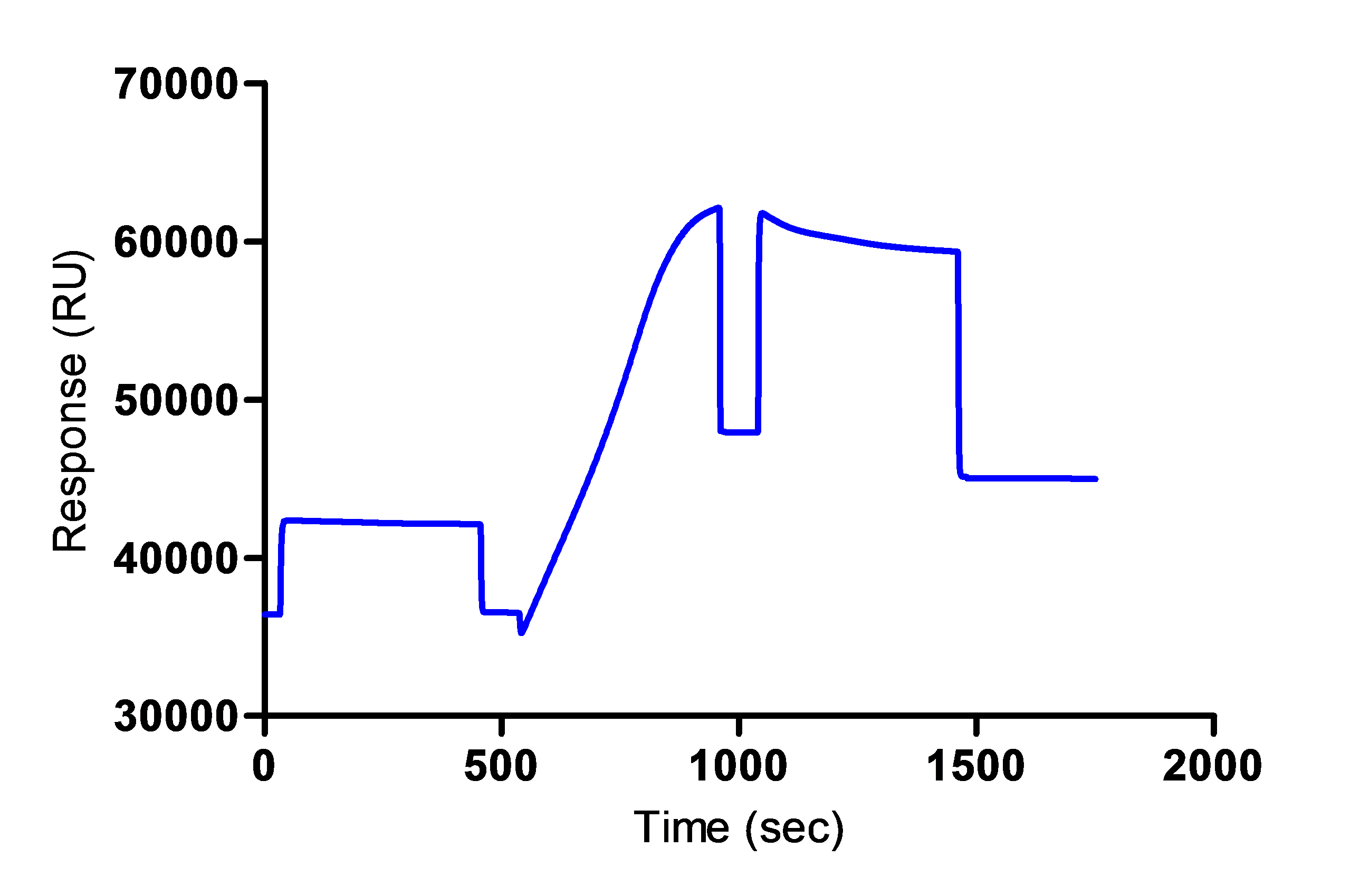
Figure 1. Immobilization of the anti-mouse IgG on the sensor chip - Prepare 300 µl of hybridoma supernatant solution 5-fold diluted by mixing 60 µl of the sample with 240 µl of 10 mM acetate buffer (pH 5.5).
- After the regeneration step, (in which the non-covalent bound anti-mouse IgG is removed from the surface without damaging non-covalent bound anti-mouse IgG), the antibodies from the hybridoma supernatants are captured non-covalently onto the surface via their Fc region by the anti-mouse IgG and the resulting complex is stabilized using a crosslink protocol independently on each flow cell.
- After antibody coupling, 3 successive injections of regeneration were done.
- The reagents and parameters used for cross-linking of the captured antibodies to the anti-mouse IgG (short activation of the carboxy groups, ligand immobilization, deactivation and regeneration) are compiled in Table 2.
Note: A maximum binding level of the antibodies coupled to the anti-mouse IgG around 1,500-2,500 RU should be obtained. Multiple injections of the diluted hybridoma supernatants can be done for reaching this binding level.
Table 2. Procedure of immobilization of the antibodies captured on the immobilized anti-mouse IgG antibodyCycle Reagent Flow cell (fc) Contact time (sec) Flow rate (µl/min) 3 1:1 (v/v) mixture of EDC (0.4 M) and NHS (0.1 M) 2, 3 or 4 30 5 µl/min 4 Hybridoma supernatant antibody sample 2, 3 or 4 600-1,800 5 µl/min 5 1 M ethanolamine-HCl (pH 8.5) 2, 3 or 4 420 5 µl/min 6 10 mM Glycine-HCl (pH 1.7) 2, 3 or 43 times 60 50 µl/min - An example of the sensorgram obtained for the antibody sample coupling is shown in the Figure 2 below.
Note: Alternatively, non-covalent capture of the antibodies without crosslinking can be performed by omitting the cycle 3 and cycle 5. For that, direct injection of the antibody sample is done after immobilization with the anti-mouse IgG, without performing an activation or deactivation of the surface. The non-covalent capture produces similar immobilization levels but the binding signal could decrease over cycles due to baseline drift during measurement and regeneration leading to lower dynamic range and decrease of the sensitivity of the assay.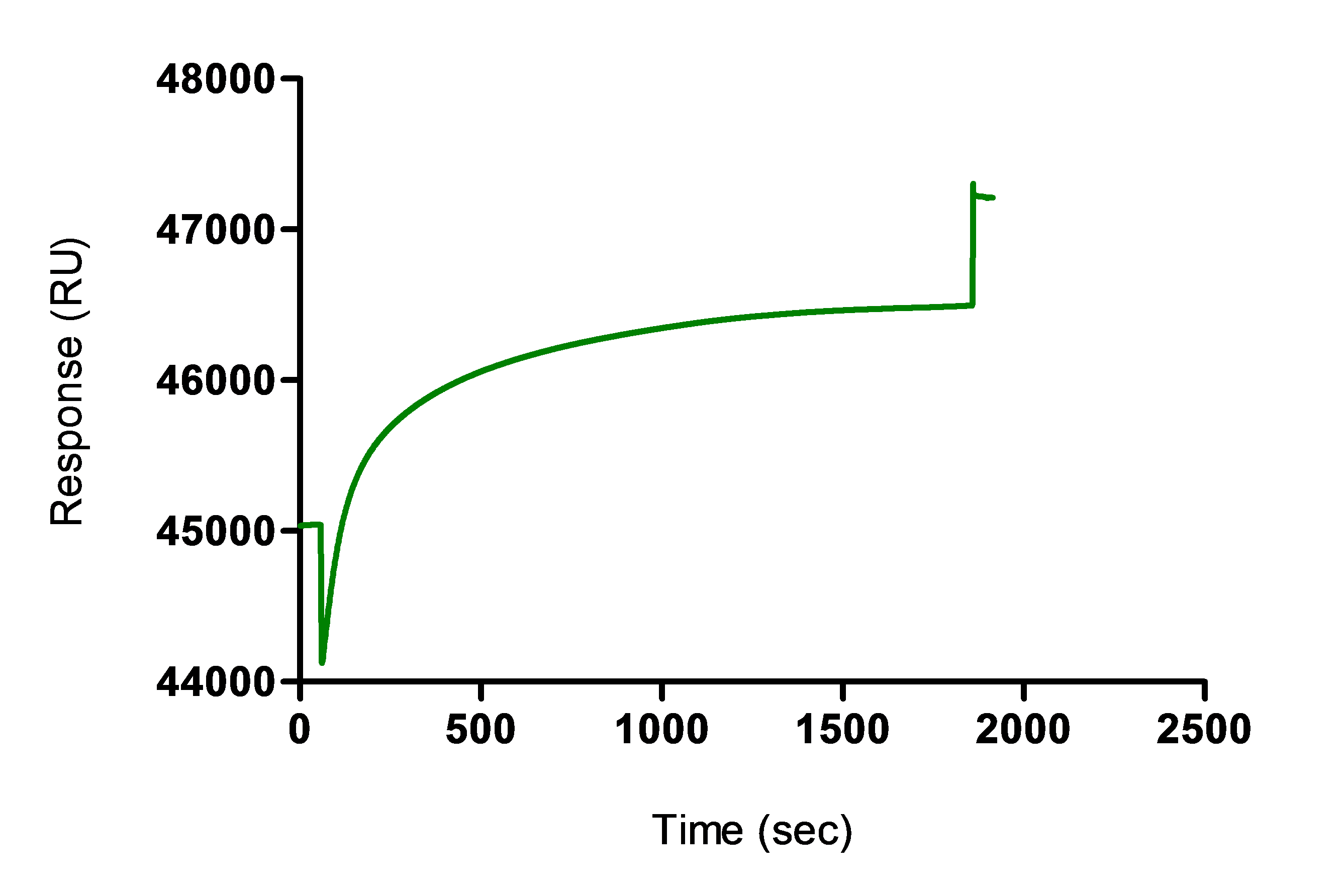
Figure 2. Coupling of antibody sample using the cross-linking protocol (cycle 4)
- Prepare 167 µl of 30 µg/ml anti-mouse IgG solution in the 10 mM acetate buffer (pH 5).
- Analyte preparation
In order to have an accurate measurement, the analyte should be in the same buffer as the continuous flow buffer to minimize bulk refractive index differences and avoid the so called bulk effect, so that can lead to low signal-to-noise-ratios. This is often most easily achieved through dilution of a concentrated analyte stock into running buffer. Therefore, the protein was diluted in the running buffer at a concentration of 4,000, 2,000, 1,000, 5,00, 250, 125, 62.5, 31.25, 15.63 and 0 nM before analysis. - Binding assay
- The binding assay was performed using PBS as running buffer.
- Each dilution of the analyte sample was injected over flow cells 1, 2, 3 and 4. Injections were performed starting from 0 (PBS buffer blank) and followed by the lowest to the highest concentration at a flow rate of 50 µl/min for 90 sec.
Once the injection was finished, the dissociation was performed by passing running buffer over the chip during 600 sec.
Note: The contact time of the association phase should be long enough to have a curvature shape on the sensorgram and the contact time of the dissociation phase should be long enough to have a flat sensorgram after dissociation phase. - After association and dissociation of the analyte, a regeneration step was performed in order to remove the remaining bound analyte. The 10 mM Glycine-HCl (pH 1.7) was selected as the optimal regeneration solutions because it allows the removal of the analyte bound to the ligand without changing the activity of the immobilized ligand. This was confirmed by the equal responses obtained from the binding assays before and after regeneration. Regeneration was performed with one injection of 3 sec (2.5 µl) of the regeneration solution.
An example of the binding assay is shown in Table 3 below.
Table 3. Summary of the binding assay conditionsNote: *Binding is studied on the sensorgrams subtracted with the reference flow cell fc1.Cycle Status Reagent Flow cell (fc)* Contact time (sec) Flow rate (µl/min) 7 Association Analyte (protein)
(0 and 15.63-4,000 nM)2-1, 3-1 or 4-1 90 50 Dissociation PBS buffer blank 2-1, 3-1 or 4-1 600 50 Regeneration 10 mM Glycine-HCl (pH 1.7) 2-1, 3-1 or 4-1 3 50
A sensorgram of protein-antibody sample interaction is shown in Figure 3.
Figure 3. Binding assay
- The binding assay was performed using PBS as running buffer.
- Data analysis
Data analysis is performed with the Biacore T100 Evaluation Software.- Binding to the reference flow cell 1 was subtracted from each sample curve and maximal response unit was measured at 90 sec post injection.
- Best fitting was then established for 5 concentration of protein using the software. In general a 1:1 model Langmuir was selected for the fitting based on the expected interaction; alternatively in some cases a two-state reaction model was chosen.
- The kinetics constants (on-rate ka and off-rate kd) as well as the affinity (in terms of equilibrium dissociation constant KD) were then calculated. An example of the final fitting results using 5 concentrations of protein and with a model 1:1 Langmuir fitting (black curves) is shown in the Figure 4 and Table 4.
Note: The final affinity measurement reported for the protein- antibody sample interaction was selected based on the Chi-square (Chi2), U-value and quality index.
The chi-square value is a quantitative measure of the closeness of fit, and in an ideal situation will approximate to the square of the short-term noise level. It is however difficult to recommend absolute values for acceptance limits for chi square: the values need to be considered from case to case, but it is advised to consider kinetics results with the lowest chi-square value.
The U-value obtained using the 1:1 binding model fitting, is an additional indicator of the parameter significance. This is a parameter that represents the uniqueness of the calculated rate constants and Rmax, determined by testing the dependence of fitting on correlated variations between selected variables. Lower values indicate greater confidence in the results. A high value (above about 25) indicates that the reported kinetic constants contain no useful information. Finally, kinetic constants should be uniquely determined and within the limitation of the instrument.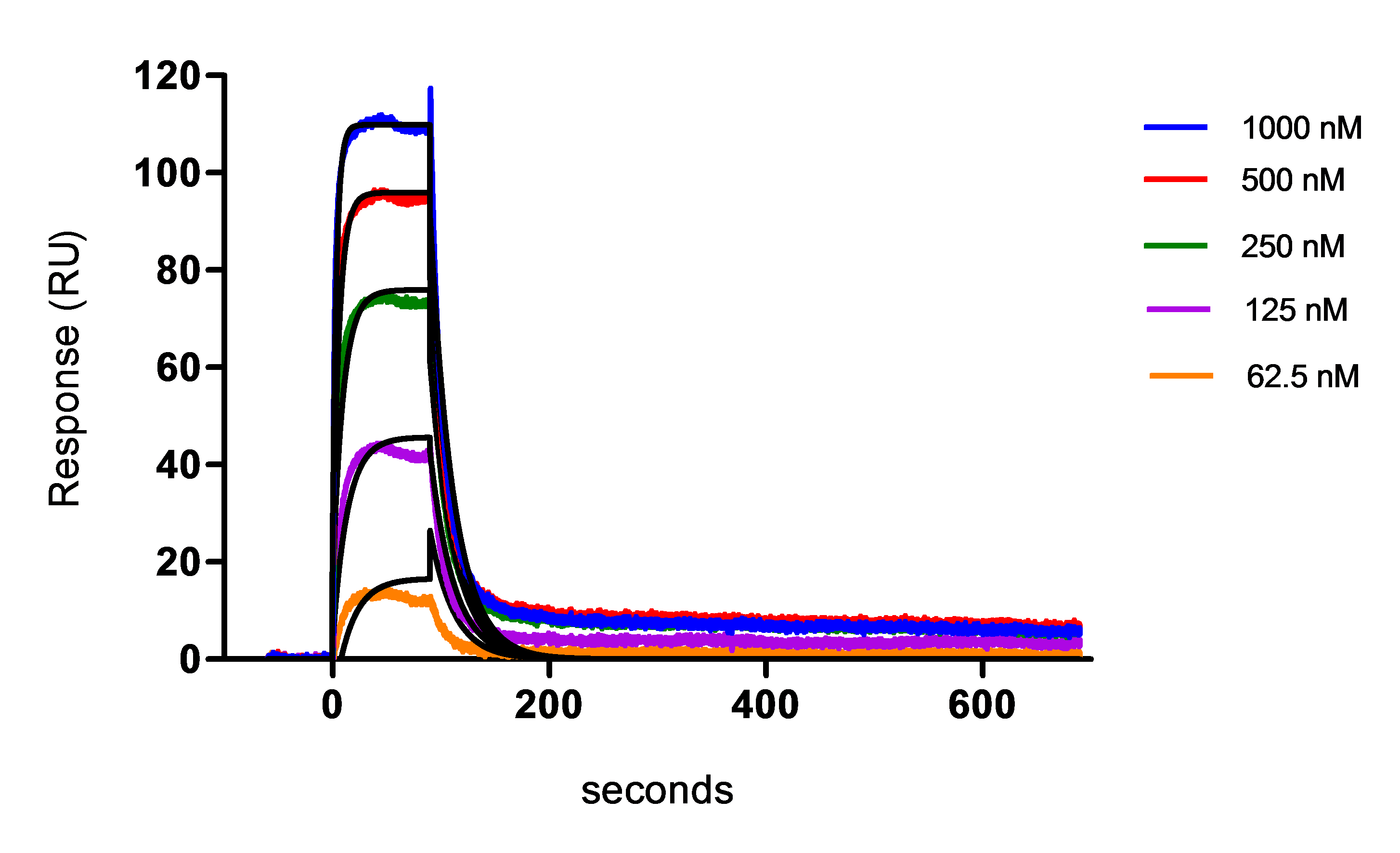
Figure 4. Affinity measurement
Table 4. Example of data extracted from the fitting procedure using the Biacore T100 evaluation software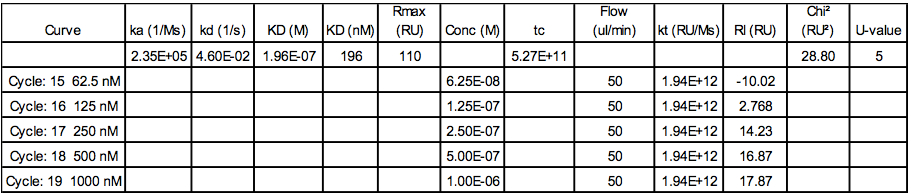
Note: Rmax corresponds to the analyte binding capacity of the surface,
tc corresponds to the flow rate independent part of the mass transfer constant,
kt corresponds to the mass transport constant and is calculate as following:
kt= km*MW*109
Where km corresponds to the mass transport coefficient and is a function of the flow rate, flow cell dimensions and diffusion properties of the analyte,
RI to the bulk refractive index contribution in the sample.
- Binding to the reference flow cell 1 was subtracted from each sample curve and maximal response unit was measured at 90 sec post injection.
Notes
Please follow the manual of your SPR instrument to implement this protocol.
In our hands, the average ± standard deviation for immobilization levels from 31 binding assay analyses was 2,326 ± 558 RUs.
Recipes
The following recipes were used to prepare the polyclonal antibody samples from our hybridoma supernatants and protein that were optimal for our assay.
- Reagents for immobilization
Running buffer, Dulbecco’s phosphate buffered saline buffer, modified without calcium chloride and magnesium chloride which contains:
2.7 mM KCl
1.5 mM KH2PO4
136.9 mM NaCl
8.9 mM Na2HPO4.7H2O - Ligands
Anti-mouse IgG (Polyclonal rabbit anti-mouse immunoglobulin) diluted in 10 mM acetate buffer (pH 5.0)
Polyclonal antibody sample from hybridoma supernatants, diluted 5 fold in 10 mM acetate buffer (pH 5.5). The standard procedure for hybridome supernantant generation is the following: After the fusion, the motherclones are plated onto 96-well plates (typically 32 plates with 50,000 cells/well) and the fusions are kept in culture for 10-14 days until clones become visible and cells are ready for screening. Four days before screening, the cell medium is changed in order to avoid false-positive signals from antibodies produced by non-surviving clones. The supernatants from wells containing surviving hybridoma cells (i.e. the splenocytes which successfully fused with myeloma cells) are tested by Luminex (for the presence of antibodies with the desired binding profiles, likely “polyclonal” response) and selected motherclones are subcloned. Serial dilutions subcloning are done in an attempt to obtain single cell per well. Cells are left to grow for about two weeks and the wells containing single clones are tested for the presence of specific monoclonal antibodies. The stable, clonal hybridoma cells are expanded for 2 weeks in the serum-free cell culture medium. The supernatants are tested by Luminex to evaluate the concentration of the mouse IgG. Typically, the concentrations varied between 10 to 65 μg/ml. The example provided in Figure 4 and Table 4 is a polyclonal supernatant of a mother clone with a concentration of 20 μg/ml. - Reagents for binding assay
Running buffer, Dulbecco’s phosphate buffered saline buffer, modified without calcium chloride or magnesium chloride which contains:
2.7 mM KCl
1.5 mM KH2PO4
136.9 mM NaCl
8.9 mM Na2HPO4.7H2O - Analyte
Purified protein diluted in the running buffer at 0.5 mg/ml
References
- Canziani, G. A., Klakamp, S. and Myszka, D. G. (2004). Kinetic screening of antibodies from crude hybridoma samples using Biacore. Anal Biochem 325 (2): 301-307.
- Schraml, M. and Biehl, M. (2012). Kinetic screening in the antibody development process. Methods Mol Biol 901: 171-181.
- Pope, M. E., Soste, M. V., Eyford, B. A., Leigh Anderson, N., Pearson, T. W. (2009). Anti-peptide antibody screening: selection of high affinity monoclonal reagents by a refined surface plasmon resonance technique. J Immunol Methods 341 (1-2): 86-96.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Ndao, D. M., Hickman, D. T., López-Deber, M. P., Davranche, A., Pfeifer, A. and Muhs, A. (2014). Binding Affinity Measurement of Antibodies from Crude Hybridoma Samples by SPR. Bio-protocol 4(21): e1276. DOI: 10.21769/BioProtoc.1276.
Category
Immunology > Antibody analysis > Antibody-antigen interaction
Immunology > Antibody analysis > Antibody function
Biochemistry > Protein > Interaction > Protein-protein interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link