Advanced Search
LC-MS/MS: the identification of methylation abundance in DNA and mRNA
Last updated date: Aug 15, 2022 Views: 576 Forks: 0
Abstract: DNA and mRNA methylation play crucial roles in controlling various biological processes and exhibit great potential in improving plant development and environmental adaptability. However, the progress in exploiting the biological function underlying DNA and mRNA modification is once restricted by technological difficulties. With the developing of liquid chromatography–tandem mass spectrometry (LC–MS/MS), novel DNA and mRNA marks are being widely identified.Here, we describe a protocol to detect the modification abundance of DNA and mRNA, which includes the detail materials, reagents, result interpretation and some cautions to avoid inaccurate and even false results. The procedure will help researchers understand and conduct the assay more conveniently.
Keywords: nucleic acid methylation, DNA methylation, RNA methylation, LC-MS/MS, nucleosides
Background
Favorable alleles for desirable traits have been integrated in crops by natural and artificial selection for millennia. However, the steady increases in crop yields over the past few years are hard to feed an incremental population on a planet currently undergoing enormous climactic changes. Novel breeding trajectories will be required to fill the predicted yield gaps. Significant progress beyond genome, especially DNA and mRNA methylation, have corroborated the crucial roles of epigenetic marks in regulating development and environmental response (Cui et al., 2017; Liang et al., 2018; Zhang et al., 2018; Zhou et al., 2018).
The progress in understanding the biological function of DNA and mRNA modification is once very rare because of technological difficulties. With recent advances in technologies such as high-throughput sequencing and liquid chromatography–tandem mass spectrometry (LC–MS/MS), DNA and mRNA modifications are being extensively characterized. The identification and genome-wide analysis of N7-methylguanine (m7G), N6-methyladenosine (m6A), N1-methyladenosine (m1A), 5-methylcytosine (m5C), 5-hydroxymethylcytosine (hm5C), 2’-O-methylated nucleosides (Nm), inosine (I), pseudouridine (J), N4-acetylcytidine (ac4C) and uridylation have been uncovered to modulate development and gene expression in eukaryotic species. Among them, m6A and m5C play key and dynamic roles in plant development and environmental adaptability (Liang et al., 2020). The function of RNA m6A has been grandly testified to involve various biological processes, including embryo development, floral transition, male meiosis, tapetal degradation, nitrate metabolism and root development (Shen et al., 2016; Duan et al., 2017; Martínez-Pérez et al., 2017; Růžička et al., 2017; Arribas-Hernández et al., 2018; Wei et al., 2018; Hou et al., 2021; Ma et al., 2021; Cheng et al., 2022). m5C modification widely occurs in Arabidopsis (~0.027%, m5C/C), Oryza sativa (Nipponbare, ~0.025%), Medicago truncatula (R108, ~0.082%), Zea mays (B73, ~0.04%), and Setaria italica (Yugu1, 0.02%), and it is proved to control root development and heat resistance (Cui et al., 2017; David et al., 2017; Yang et al., 2019; Tang et al., 2020). DNA N6-methyldeoxyadenosine (6mA), another new epigenetic mark, has been recently identified during plant development and stress responses, and DNA 6mA level is changing from 0.0264% (6mA/A) in soybean, 0.35% in sorghum, 0.048% in Arabidopsis, and 0.55% in rice based on the LC-MS/MS results (Liang et al., 2018; Zhang et al., 2018; Zhou et al., 2018; Ye et al., 2019; Yuan et al., 2020).
In LC–MS/MS assay to detect the methylation abundance, genomic DNA and mRNA is digested into single nucleosides and dephosphorylated. Individual nucleosides are resolved and then subjected to triple quadrupole mass spectrometry. Nucleosides are quantified using the nucleoside-to-base ion mass transitions (Liang et al., 2018). This protocol shows readers how to conduct LC–MS/MS analysis to detect genomic DNA and mRNA methylation.
Materials, equipment and Reagents
1. Materials and equipment
(1) Centrifugal Filter Units (Amicon Ultra, 10K MWCO)
(2) Vial & screw (Agilent Technologies)
(3) Hypersil GOLD a Q reverse phase column (Thermo Scientific)
(4) Agilent 6490 Triple Quadrupole mass spectrometer
2. Reagents
(1) EHNA (Sigma-Aldrich, E114-25MG)
(2) Desferrioxamine (Sigma-Aldrich, D9533-1G)
(3) Tetrahydrouridine (abcom, ab142080-10mg)
(4) Butylated hydroxytoluene (Sigma-Aldrich, B1215000)
(5) Nuclease P1 (Sigma-Aldrich, N8630-1VL)
(6) Phosphodiesterase I (Sigma-Aldrich, P3134-100MG)
(7) Sodium acetate (Sigma-Aldrich, S2889)
(8) Zinc chloride (Sigma-Aldrich, 208086)
(9) DNA extraction kit (TIANGEN, DP350-02)
(10) RNA Easy Fast (TIANGEN, DP452)
(11) A standard
(12) 6mA standard
Procedure
1. Isolation of nucleic acid
The genomic DNA and total RNA are extracted from plants using plant genomic DNA extraction kit (TIANGEN, DP350-02) and RNA Easy Fast extraction kit (TIANGEN, DP452) respectively according to the manufacturer’s instructions. Note: the RNA in isolated genomic DNA samples and DNA in total RNA should be eliminated in case interfering the quantification of DNA and RNA methylation. The purity of total RNA can be interrogated using Agilent 2100 Bioanalyzer Plant RNA Pico assay.
2. Digestion and purification of nucleic acid
0.5~1 μg genomic DNA samples are digested into single nucleosides in a digestion buffer (Table 1) for ~5 h at 37℃. Similarly, 0.5~1 μg RNA samples are usually digested for ~3 h. Subsequently these single nucleosides are dephosphorylated with bacterial alkaline phosphatase (10 U) for 1 h at 37℃. Enzymes in digestion buffer are removed by filtration (Amicon Ultra 10K MWCO) at 16000 g for 5 min.
Note: deaminase inhibitors (coformycin, tetrahydrouridine) and antioxidants (desferrioxamine, butylated hydroxytoluene) should be added to samples prior to digestion. Nucleic acid samples are digested into single ribonucleosides by applying nuclease P1 and phosphodiesterase I.
Table 1 Nucleic acid digestion buffer
Components | Final concentration |
Coformycin | 0.005 μg/μl |
Desferrioxamine | 0.1 mM |
Tetrahydrouridine | 0.05 μg/μl |
Butylated hydroxytoluene | 0.1 mM |
Nuclease P1 | 1U |
Phosphodiesterase I | 0.25 mg/μl, 0.01 U |
Sodium acetate (pH 5.2) | 30 mM |
Zinc chloride | 2 mM |
3. LC-MS/MS analysis
Single nucleosides are resolved on a Hypersil GOLD a Q reverse phase column (Thermo Scientific, 100×2.1 mm, 1.9 μm) eluted with a gradient of acetonitrile/0.1% formic acid at flow rate of 0.3 ml/min and 25℃: 0-15.3 min, 0-1%; 15.3-20 min, 1-6%; 20-30 min, 6-100%, 30-41 min, 100-0%. Samples are then subjected to LC-MS/MS analysis on an Agilent 6490 Triple Quadrupole mass spectrometer operated in positive ion mode with the following source parameters: gas temperature, 50℃; gas flow, 11 L/min; nebulizer, 20 psi; sheath gas temperature, 300℃; sheath gas flow, 12 L/min and capillary voltage 1800 V.
Result interpretation
Various nucleic acid methylation types are distinguished according to chromatographic retention times and unique m/z of commercially available synthetic standards. For instance, nucleosides are quantified using the nucleoside-to-base ion mass transitions of 266.1 to 150.1 for 6mA and 252.1 to 136.1 for dA, which is the criterion to determine different chemical modifications. The retention times and m/z of the parent ion and product ion are shown as Table 2.
Table 2 The common parent ion, product ion of ribonucleosides and retention times
Compound Name | Precursor Ion (unit) | Product Ion (unit) |
m6dA | 266.1 | 150.1 |
dA | 252.1 | 136.1 |
4/5dC | 242.2 | 126.1 |
dC | 228.2 | 112.1 |
m6A | 282.1 | 150.1 |
rA | 268.1 | 136.1 |
m5C | 258.1 | 126.1 |
rC | 244.1 | 112.1 |
We select DNA 6mA as a sample to exhibit the LC-MS/MS data. The absolute level of DNA 6mA and dA in each sample can be quantified by DNA 6mA and dA standard substance, by which the mass spectrometer signal intensities of DNA 6mA are transformed into concentration (Fig. 1A, B). Subsequently the relative abundance of DNA 6mA in each sample is determined by the concentration of 6mA/∑(dA, 6mA) (Fig. 1C, D).
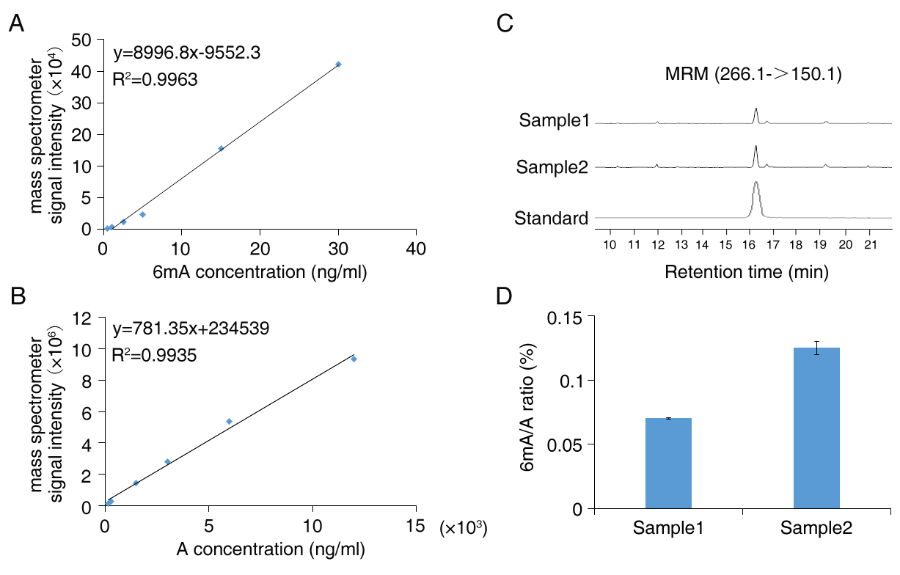
Figure 1 LC-MS/MS quantification of DNA 6mA levels。 (A-B) the standard curve of DNA 6mA and A. (C) Ion chromatograms of DNA 6mA levels. (D) Quantification of the DNA 6mA abundance relative to adenine (6mA/A ratio).
Discussion
LC-MS/MS is an accurate method in detecting the absolute methylation abundance in genomic DNA and mRNA. Many studies also launched dot blotting assay with specific antibody to test the relative methylation level in nucleic acid. However, this means can not determine the absolute methylation level, and sometimes the results of dot blotting are fluctuant. Several high-throughput sequencing (HTS) based technologies are also used to identify nucleic acid methylation sites. Antibody-based HTS, such as 6mA IP-seq, is performed to detect 6mA peaks at ~100 bp resolution (Liang et al., 2018). Single molecule real-time (SMRT) sequencing can identify 6mA at single-base resolution by means of unique kinetic signatures for each modification (Luo et al., 2015). LC-MS/MS and high-throughput sequencing usually validate the results of each other. In our previous studies, the 6mA level in various tissues and rice subspecies tested by LC-MS/MS are coincident with the results of 6mA IP-seq and SMRT-seq (Liang et al., 2018; Zhang et al., 2018). To date, LC-MS/MS has been extensively utilized to detect various modifications in plants, such as Arabidopsis, rice, maize, soybean, Casuarina equisetifolia (Cui et al., 2017; Liang et al., 2018; Zhang et al., 2018; Zhou et al., 2018; Ye et al., 2019; Yuan et al., 2020). This protocol provides coherent procedures and some cautions to avoid inaccurate and even false results. Readers can use this protocol to check DNA and mRNA modification in samples of interest.
Acknowledgments
This protocol was supported by the National Natural Science Foundation of China (32130080, 31871606, 32101786), the Central Public-Interest Scientific Institution Basal Research Fund (Y2020PT06, Y2022PT22), and Beijing Natural Science Foundation (6222055).
Competing interests
The authors declare that there are no conflicts of interest or competing interests.
References
Arribas-Hernández, L., Bressendorff, S., Hansen, M.H., Poulsen. C., Erdmann, S., and Brodersen, P. (2018). An m6A-YTH module controls developmental timing and morphogenesis in Arabidopsis. Plant Cell 30, 952–967.
Cui, X., Liang, Z., Shen, L., Zhang, Q., Bao, S., Geng, Y., Zhang, B., Leo, V., Vardy, L.A., Lu, T., et al. (2017). 5-methylcytosine RNA methylation in Arabidopsis thaliana. Mol Plant 10, 1387–1399.
Cheng, P., Bao, S., Li, C., Tong, J., Shen, L., and Yu, H. (2022). RNA N6-methyladenosine modification promotes auxin biosynthesis required for male meiosis in rice. Dev Cell 57, 246–259.
David, R., Burgess, A., Parker, B., Li, J., Pulsford, K., Sibbritt, T., Preiss, T., and Searle, I.R. (2017). Transcriptome-wide mapping of RNA 5-methylcytosine in arabidopsis mRNAs and noncoding RNAs. Plant Cell 29, 445–460.
Duan, H.C., Wei, L.H., Zhang, C., Wang, Y., Chen,. L, Lu, Z., Chen, P.R., He, C., and Jia, G. (2017). ALKBH10B is an RNA N6-Methyladenosine demethylase affecting Arabidopsis floral transition. Plant Cell 29, 2995–3011.
Hou, Y., Sun, J., Wu, B., Gao, Y., Nie, H., Nie, Z., Quan, S., Wang, Y., Cao, X., and Li, S. (2021). CPSF30-L-mediated recognition of mRNA m6A modification controls alternative polyadenylation of nitrate signaling-related gene transcripts in Arabidopsis. Mol Plant 14, 688–699.
Luo, G.Z., Blanco, M.A., Greer, E.L., He, C., and Shi, Y. (2015). DNA N6-methyladenine: a new epigenetic mark in eukaryotes? Nat Rev Mol Cell Biol 16, 705-710.
Liang Z, Riaz A, Chachar S, Ding Y, Du H, Gu X. (2020). Epigenetic modifications of mRNA and DNA in plants. Mol Plant 13(1), 14-30.
Liang, Z., Shen, L., Cui, X., Bao, S., Geng, Y., Yu, G., Liang, F., Xie, S., Lu, T., Gu, X., and Yu, Hao. (2018). DNA N6 -Adenine Methylation in Arabidopsis thaliana. Dev Cell 45, 406-416.
Liang, Z., Zhang, Q., Ji, C., Hu, G., Zhang, P., Wang, Y., Yang, L., and Gu, X.. (2021). Reorganization of the 3D chromatin architecture of rice genomes during heat stress. BMC Biol 19, 53.
Ma, K., Han, J., Zhang, Z., Li, H., Zhao, Y., Zhu, Q., Xie, Y., Liu, Y., and Chen, L. (2021). OsEDM2L mediates m6A of EAT1 transcript for proper alternative splicing and polyadenylation regulating rice tapetal degradation. J Integr Plant Biol 63, 1982-1994.
Martínez-Pérez, M., Aparicio, F., López-Gresa, M.P., Bellés,. JM., Sánchez-Navarro, J..A, and Pallás, V. (2017). Arabidopsis m6A demethylase activity modulates viral infection of a plant virus and the m6A abundance in its genomic RNAs. Proc Natl Acad Sci USA 114, 10755–10760.
Růžička, K., Zhang, M., Campilho, A., Bodi, Z., Kashif, M., Saleh, M., Eeckhout, D., El-Showk. S., Li, H., Zhong, S., et al. (2017). Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phyto 215, 157–172.
Shen, L., Liang, Z., Gu, X., Chen, Y., Teo, Z.W.N., Hou, X., Cai, W.M., Dedon, P..C, Liu, L., and Yu, H. (2016). N6-Methyladenosine RNA Modification Regulates Shoot Stem Cell Fate in Arabidopsis. Dev Cell 38, 186–200.
Tang, Y., Gao, C., Gao, Y., Yang, Y., Shi, B., Yu, J.L., Lyu, C., Sun, B., Wang, H.L., Xu, Y., Yang, Y., Chong, K. (2020). OsNSUN2-mediated 5-methylcytosine mRNA modification enhances rice adaptation to high temperature. Dev Cell 53, 272–286.
Wei, L.H., Song, P., Wang, Y., Lu, Z., Tang, Q., Yu, Q., Xiao, Y., Zhang, X., Duan, H.C., Jia, G. (2018). The m6A reader ECT2 controls trichome morphology by affecting mRNA stability in Arabidopsis. Plant Cell 30, 968–985.
Yang, L., Perrera, V., Saplaoura. E., Apelt, F., Bahin, M., Kramdi, A., Olas, J., Mueller-Roeber, B., Sokolowska, E., Zhang, W., Li, R., Pitzalis, N., Heinlein, M., Zhang, S., Genovesio, A., Colot, V., and Kragler, F. (2019). m5C methylation guides systemic transport of messenger RNA over graft junctions in plants. Curr Biol 29, 2465–2476.
Ye, G., Zhang, H., Chen, B., Nie, S., Liu, H., Gao, W., Wang, H., Gao, Y., and Gu, L. (2019). De novo genome assembly of the stress tolerant forest species Casuarina equisetifolia provides insight into secondary growth. Plant J 97, 779-794.
Yu Q, Liu S, Yu L, Xiao Y, Zhang S, Wang X, Xu Y, Yu H, Li Y, Yang J, et al. 2021. RNA demethylation increases the yield and biomass of rice and potato plants in field trials. Nat Biotech 39(12):1581-1588.
Yuan, D., Xing, J., Luan, M., Ji, K., Guo, J., Xie, S., and Zhang, Y. (2020). DNA N6-methyladenine modification in wild and cultivated soybeans reveals different patterns in nucleus and cytoplasm. Front Genet 736.
Zhang, Q., Liang, Z., Cui, X., Ji, C., Li ,Y., Zhang, P., Liu, J., Riaz, A., Yao, P., Liu,. M., et al. (2018). N6-Methyladenine DNA methylation in Japonica and Indica rice genomes and its association with gene expression, plant development, and stress responses. Mol Plant 11, 1492–1508.
Zhou, C., Wang, C., Liu, H., Zhou, Q., Liu, Q., Guo, Y., Peng, T., Song, J., Zhang, J., Chen, L., et al. (2018). Identification and analysis of adenine N6-methylation sites in the rice genome. Nat Plants 4, 554–563.
- yang, L, Li, X, Zhang, Q and Gu, X(2022). LC-MS/MS: the identification of methylation abundance in DNA and mRNA. Bio-protocol Preprint. bio-protocol.org/prep1861.
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
