- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Arginine-rich Peptides Can Actively Mediate Liquid-liquid Phase Separation
Published: Vol 7, Iss 17, Sep 5, 2017 DOI: 10.21769/BioProtoc.2525 Views: 14224
Reviewed by: Nicoletta CordaniWeiyan JiaMirko Messa

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation and Imaging of Microvessels From Brain Tissue
Josephine K. Buff [...] Sophia M. Shi
Aug 5, 2025 2631 Views

Protocol for the Preparation of a Recombinant Treacle Fragment for Liquid–Liquid Phase Separation (LLPS) Assays
Nadezhda V. Petrova [...] Artem K. Velichko
Sep 20, 2025 1820 Views

Optimized Secretome Sample Preparation From High Volume Cell Culture Media for LC–MS/MS Proteomic Analysis
Basil Baby Mattamana [...] Peter Allen Faull
Dec 20, 2025 1246 Views
Abstract
Studying liquid-liquid phase separation (LLPS) of proteins provides key insights into the biogenesis of membraneless organelles and pathological protein aggregation in disease. We have established a protocol for inducing the phase separation of arginine-rich peptides, which allows for studying their molecular determinants and dynamics (Boeynaems et al., 2017).
Keywords: LLPSBackground
Arginine-rich disordered domains are often found in RNA binding proteins, including the ones associated with neurodegenerative diseases (e.g., FUS, FMRP, hnRNPA1) (Varadi et al., 2015; Boeynaems et al., 2017). Also toxic arginine-rich repeat peptides (i.e., polyGR and polyPR) are produced in amyotrophic lateral sclerosis patients carrying the C9orf72 repeat expansion (Kwon et al., 2014; Mizielinska et al., 2014; Varadi et al., 2015; Boeynaems et al., 2016). While phase separation of uncharged low complexity domains had been studied before (Kato et al., 2012; Burke et al., 2015; Lin et al., 2015; Molliex et al., 2015; Patel et al., 2015), we developed this protocol to test whether arginine-rich domains could also contribute to phase separation. A schematic representation of the protocol can be seen in Figure 1.
Figure 1. Workflow of protein droplet formation and analysis. A diffuse arginine-rich peptide solution can be induced to undergo liquid-liquid phase separation by addition of RNA or molecular crowder (PEG). Resulting droplets can be further analyzed by fluorescence microscopy and FRAP.
Materials and Reagents
- Pipette tips
- Amicon® Ultra–0.5 ml centrifugal filters 3K (Merck, catalog number: UFC500396 )
- Cell counting slides (Bio-Rad Laboratories, catalog number: 1450015 )
- Clear adhesive tape
- Custom 20 to 60 amino acid peptides with free carboxy and amino-termini (Pepscan)
- Custom Alexa labeled RNA 30 ribonucleotide oligomers (IDT)
- MilliQ water
- Alexa Fluor® Protein Labeling Kit (Thermo Fisher Scientific, InvitrogenTM, catalog number: A10235 )
- PEG300 (Sigma-Aldrich, catalog number: 81160 )
Note: This product has been discontinued. - Potassium phosphate monobasic (KH2PO4)
- Potassium phosphate dibasic (K2HPO4)
- Polyuridylic acid potassium salt (Sigma-Aldrich, catalog number: P9528 )
- Clear nail varnish
- 10x potassium buffer, pH 7 (see Recipes)
Equipment
- Pipette
- Bench-top centrifuge (Eppendorf, models: 5430 and 5810 R )
- trUView cuvettes (Bio-Rad Laboratories, catalog number: 1702510 )
Note: This product has been discontinued. - SmartSpec Plus spectrophotometer (Bio-Rad Laboratories, model: SmartSpec Plus )
- LSM 780 Meta NLO confocal microscope (ZEISS, model: LSM 780 NLO ) with 20x long range objective (ZEISS, model: LD Plan-Neofluor 20x/0.4 Corr Ph2 M27, catalog number: 421351-9970-000 )
Software
- Zen software (ZEISS)
- GraphPad Prism (GraphPad)
- Excel (Microsoft)
Procedure
Note: Dissolve aliquots of lyophilized custom peptides (Pepscan) in MilliQ water to a stock solution of 1 mM. Dissolve aliquots of lyophilized custom fluorescent RNA oligos (IDT) in MilliQ water to a stock solution of 100 µM. Dissolve lyophilized polyU potassium salt (Sigma-Aldrich) in MilliQ water to a stock solution of 10 µg/µl. Aliquots are stored at -20 °C.
- Fluorescent labeling of custom peptides
- Perform labeling reaction with desired Alexa Fluor® labeling kit according to the manufacturer’s instructions, but purify the peptide with the use of Amicon Ultra spin columns (minimum peptide size must be above 3 kDa).
- Add labeling reaction to spin column and centrifuge in bench top centrifuge for 10 min at 20,817 x g (rcf, max speed) at room temperature (Eppendorf 5430).
- Discard flow-through. Add 400 µl MilliQ water and centrifuge again. Repeat at least five times or until there is visually no more dye in the filtrate.
- Pipet remaining solution which still contains labeled peptide from spin column and adjust volume to obtain a 500 µM-1 mM stock solution. Concentration is calculated based on initial input.
- Perform labeling reaction with desired Alexa Fluor® labeling kit according to the manufacturer’s instructions, but purify the peptide with the use of Amicon Ultra spin columns (minimum peptide size must be above 3 kDa).
- Induction of arginine-peptide phase separation by molecular crowder
- Mix by pipetting rigorously together the following reagents in MilliQ water to obtain described final concentrations.
Note: Make sure to pipet PEG300 slowly and always with a new pipet tip to prevent errors due to the high viscosity of the solution.
Potassium buffer (10x stock) to 1x (see Recipes)
PEG300 (100%) to 30%
Peptide (1 mM) to desired concentration (1-250 µM) - If the peptide is prone to phase separation the solution should turn cloudy or opaque instantaneously at room temperature. For example, see Figure 2A. Briefly cooling on ice can promote phase separation of proteins that phase separate only weakly.
- Quantify extent of phase separation by measuring OD600 of 60 µl samples using trUView microcuvettes in a SmartSpec Plus (or similar) spectrophotometer.
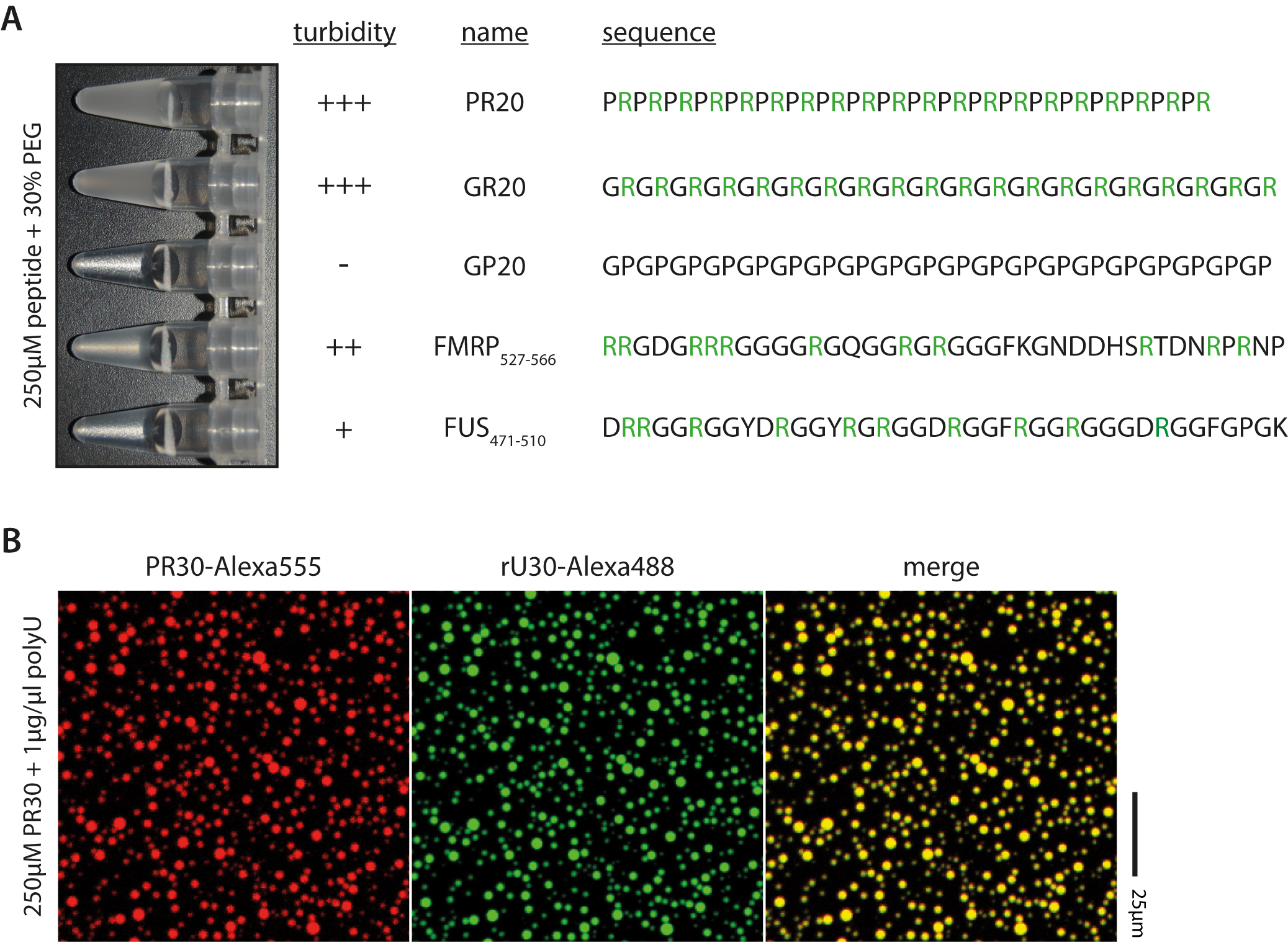
Figure 2. Examples of peptides and droplet formation. A. Peptide sequence and arginine content affect phase transition; Arginines highlighted in green. B. RNA and arginine-rich peptides colocalize upon phase separation, as seen by fluorescence microscopy.
- Mix by pipetting rigorously together the following reagents in MilliQ water to obtain described final concentrations.
- Induction of arginine-peptide phase separation by RNA
- Mix by pipetting rigorously together the following reagents in MilliQ water to obtain described final concentrations:
Potassium buffer (10x stock) to 1x
PolyU potassium salt (10 µg/µl) to 1 µg/µl
Peptide (1 mM) to desired concentration (1-250 µM) - If the peptide is prone to phase separation the solution should turn cloudy or opaque instantaneously at room temperature.
- Quantify extent of phase separation by measuring OD600 of 60 µl samples using trUView microcuvettes in a SmartSpec Plus (or similar) spectrophotometer.
- Mix by pipetting rigorously together the following reagents in MilliQ water to obtain described final concentrations:
- Imaging arginine-peptide phase separation by fluorescence microscopy
- Prepare phase separated peptide solution as described above. Spike the sample with fluorescent peptide or RNA oligomer to reach 200 nM and 100 nM respectively as final concentrations.
- Pipet 20 µl of the sample into each chamber of the cell counting slides (see Figure 3).
- Seal the chambers using clear adhesive tape and cover with nail varnish to prevent evaporation during imaging (see Figure 3).
- Let the samples equilibrate at room temperature in the dark for at least 30 min.
- Image samples on a LSM 780 Meta NLO confocal microscope with 20x long range objective. For example, see Figure 2B.
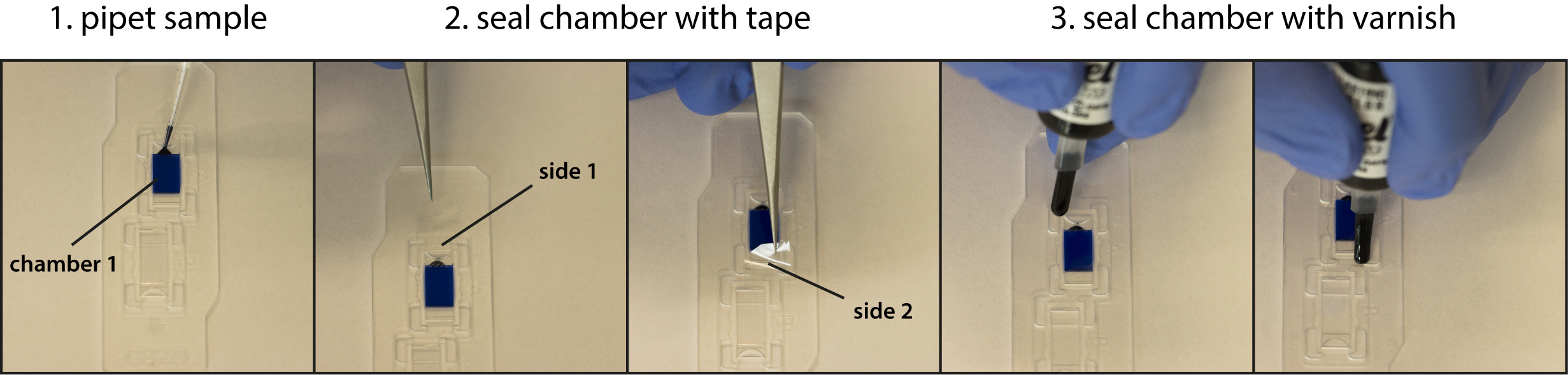
Figure 3. Workflow of microscope chamber preparation. A solution containing a blue dye was used for illustration. Step 1: Pipet sample in the incubation chamber; Step 2: Seal both sides of the chamber with clear adhesive tape; Step 3: Apply clear nail varnish to make chamber securely air tight.
- Prepare phase separated peptide solution as described above. Spike the sample with fluorescent peptide or RNA oligomer to reach 200 nM and 100 nM respectively as final concentrations.
- Fluorescence recovery after photobleaching (FRAP) analysis
- Samples are prepared as described above.
- Bleach a circular area of 1-2 µM radius at 100% laser power in the center of a droplet with a radius between 5 µM and 10 µM. Use of larger droplets is advised so bleached area does not cover whole droplet compromising the study of intradroplet diffusion. When droplets do not reach this size spontaneously, incubation chambers were briefly centrifuged for 30 sec at 201 x g (rcf; 1,000 rpm on Eppendorf 5810 R centrifuge).
- Monitor fluorescence recovery after bleaching for at least 60 sec using Zen software (Figure 4A).
- Make sure to record simultaneously the fluorescence intensity over time of the background solution and an unbleached reference droplet.
- Export raw data to Microsoft Excel for analysis.
- Samples are prepared as described above.
Data analysis
FRAP data analysis using Microsoft Excel and Prism (Figure 4B).
- Import the raw data from the Zen software into Microsoft Excel.
- Subtract the fluorescence intensity of the background from the bleached droplet and reference droplet.
- Subtract the fluorescence intensity of the first time point post-bleach from the bleached droplet. This step will normalize the first post-bleach time point to zero.
- Divide the fluorescence intensities of the bleached droplet by the pre-bleach fluorescence intensity. Do this as well for the reference droplet. This will normalize the pre-bleach time point to 1.
- Divide the fluorescence intensities of the bleached droplet by the fluorescence intensities of the reference droplet. This will correct for photobleaching due to prolonged exposure during the time course experiment.
- Import the normalized data into GraphPad Prism.
- Plot the fluorescence intensities as a function of time.
- Average FRAP curves of at least 15 droplets and evaluate differences between multiple conditions using repeated measures ANOVA.
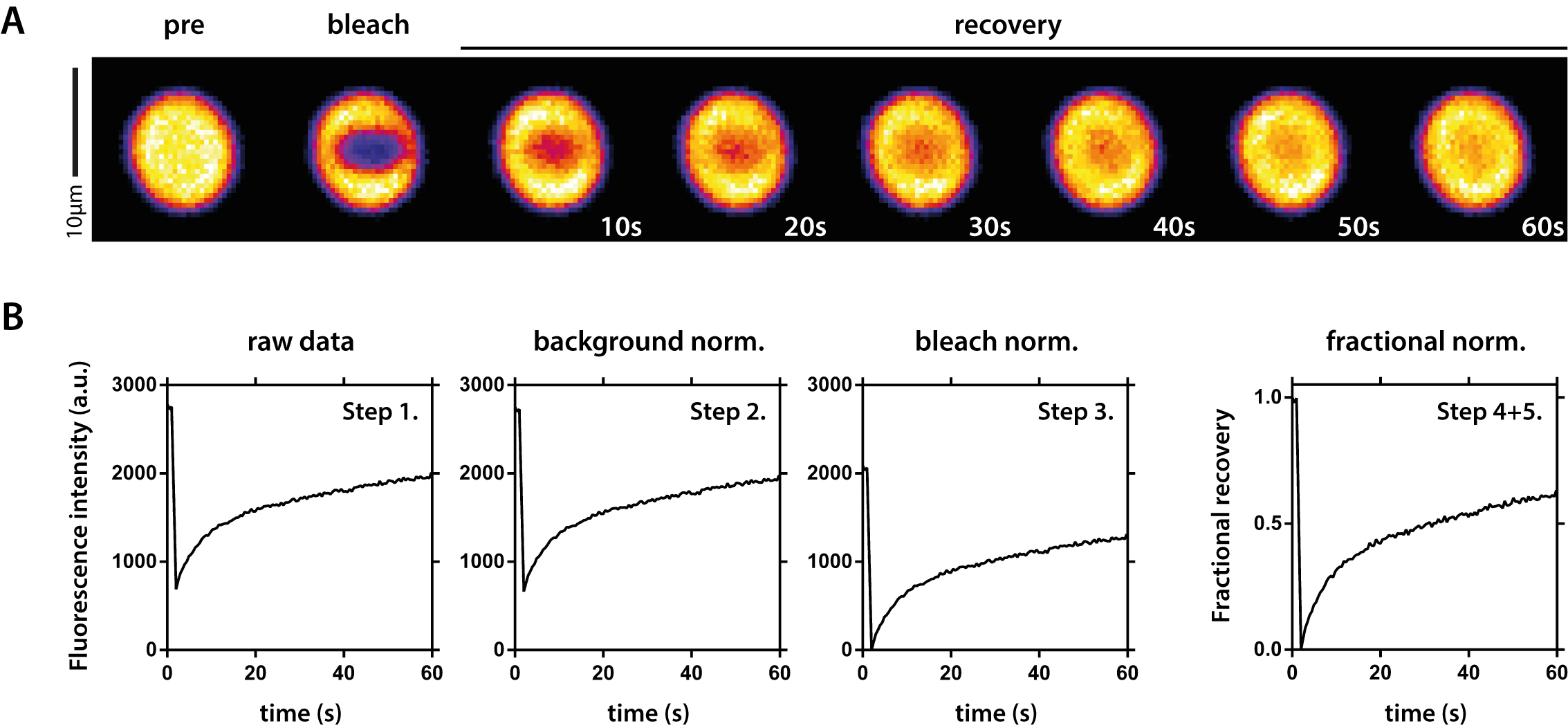
Figure 4. FRAP analysis of fluorescent protein droplets. A. Example time series of a FRAP measurement. The fluorescent PR signal was bleached inside the droplet and recovery was followed over time. B. Overview of raw data normalization. Step 1: Raw data as exported from Zen software; Step 2: Data after background normalization; Step 3: Data after bleach normalization; Step 4 + 5: Final data after fractional normalization and spontaneous bleach correction. Step numbers correspond to numbers in the text (Data analysis).
Recipes
- 10x potassium buffer
- Make 1 M stock solutions of K2HPO4 and KH2PO4 in MilliQ water
- Combine 61.5 ml of 1 M K2HPO4 with 38.5 ml of 1 M KH2PO4 to 100 ml 10x potassium buffer
- Make 1 M stock solutions of K2HPO4 and KH2PO4 in MilliQ water
Acknowledgments
This protocol was described in brief in Boeynaems et al. (2017). Research was funded by the KU Leuven, VIB, the European Research Council in the context of the European’s Seventh Framework Programme (FP7/2007-2013 and ERC grant agreement No. 340429), the Research Foundation Flanders (FWO) G.0983.14N, the Interuniversity Attraction Poles Programme P7/16 initiated by the Belgian Science Policy Office, the Association Belge contre les Maladies Neuro-Musculaires (ABMM), the ALS Liga (Belgium) and the ‘Opening the Future’ Fund. S.B. received a PhD fellowship from the Agency for Innovation by Science and Technology (IWT). P.T. was supported by the Odysseus grant G.0029.12 from Research Foundation Flanders (FWO).
References
- Boeynaems, S., Bogaert, E., Kovacs, D., Konijnenberg, A., Timmerman, E., Volkov, A., Guharoy, M., De Decker, M., Jaspers, T., Ryan, V. H., Janke, A. M., Baatsen, P., Vercruysse, T., Kolaitis, R. M., Daelemans, D., Taylor, J. P., Kedersha, N., Anderson, P., Impens, F., Sobott, F., Schymkowitz, J., Rousseau, F., Fawzi, N. L., Robberecht, W., Van Damme, P., Tompa, P. and Van Den Bosch, L. (2017). Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol Cell 65(6): 1044-1055 e1045.
- Boeynaems, S., Bogaert, E., Michiels, E., Gijselinck, I., Sieben, A., Jovicic, A., De Baets, G., Scheveneels, W., Steyaert, J., Cuijt, I., Verstrepen, K. J., Callaerts, P., Rousseau, F., Schymkowitz, J., Cruts, M., Van Broeckhoven, C., Van Damme, P., Gitler, A. D., Robberecht, W. and Van Den Bosch, L. (2016). Drosophila screen connects nuclear transport genes to DPR pathology in c9ALS/FTD. Sci Rep 6: 20877.
- Burke, K. A., Janke, A. M., Rhine, C. L. and Fawzi, N. L. (2015). Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol Cell 60(2): 231-241.
- Jovicic, A., Mertens, J., Boeynaems, S., Bogaert, E., Chai, N., Yamada, S. B., Paul, J. W., 3rd, Sun, S., Herdy, J. R., Bieri, G., Kramer, N. J., Gage, F. H., Van Den Bosch, L., Robberecht, W. and Gitler, A. D. (2015). Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci 18(9): 1226-1229.
- Kato, M., Han, T. W., Xie, S., Shi, K., Du, X., Wu, L. C., Mirzaei, H., Goldsmith, E. J., Longgood, J., Pei, J., Grishin, N. V., Frantz, D. E., Schneider, J. W., Chen, S., Li, L., Sawaya, M. R., Eisenberg, D., Tycko, R. and McKnight, S. L. (2012). Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149(4): 753-767.
- Kwon, I., Xiang, S., Kato, M., Wu, L., Theodoropoulos, P., Wang, T., Kim, J., Yun, J., Xie, Y. and McKnight, S. L. (2014). Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 345(6201): 1139-1145.
- Lin, Y., Protter, D. S., Rosen, M. K. and Parker, R. (2015). Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol Cell 60: 208-219.
- Mizielinska, S., Gronke, S., Niccoli, T., Ridler, C. E., Clayton, E. L., Devoy, A., Moens, T., Norona, F. E., Woollacott, I. O., Pietrzyk, J., Cleverley, K., Nicoll, A. J., Pickering-Brown, S., Dols, J., Cabecinha, M., Hendrich, O., Fratta, P., Fisher, E. M., Partridge, L. and Isaacs, A. M. (2014). C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 345(6201): 1192-1194.
- Molliex, A., Temirov, J., Lee, J., Coughlin, M., Kanagaraj, A. P., Kim, H. J., Mittag, T. and Taylor, J. P. (2015). Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163(1): 123-133.
- Patel, A., Lee, H. O., Jawerth, L., Maharana, S., Jahnel, M., Hein, M. Y., Stoynov, S., Mahamid, J., Saha, S., Franzmann, T. M., Pozniakovski, A., Poser, I., Maghelli, N., Royer, L. A., Weigert, M., Myers, E. W., Grill, S., Drechsel, D., Hyman, A. A. and Alberti, S. (2015). A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162(5): 1066-1077.
- Varadi, M., Zsolyomi, F., Guharoy, M. and Tompa, P. (2015). Functional advantages of conserved intrinsic disorder in RNA-binding proteins. PLoS One 10(10): e0139731.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Boeynaems, S., De Decker, M., Tompa, P. and Van Den Bosch, L. (2017). Arginine-rich Peptides Can Actively Mediate Liquid-liquid Phase Separation. Bio-protocol 7(17): e2525. DOI: 10.21769/BioProtoc.2525.
Category
Neuroscience > Nervous system disorders > Blood brain barrier
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










